METATAC V.1
Honggui Wu, Xiang Li, Fanchong Jian, Ayijiang Yisimayi, X. Sunney Xie
Abstract
Abstract
Here we describe a protocol for multiplexed end-tagging amplification of transposase accessible chromatin (METATAC), a high-sensitivity single-cell ATAC-seq technique with the help of META chemistry and extensive biochemical modifications. We improved the protocol from three aspects, first, we used Omni-ATAC protocol to permeabilize cells, which greatly reduced mitochondrial reads to less than 0.5%. Second, we used sodium dodecyl sulfate (SDS) to release Tn5 from bound DNA, which enabled maximum DNA recovery. Third, we use META transposome instead Nextera transposome used in other protocols, which avoids half loss due to self-looping during amplification, thus further increasing library size. Specifically, META sequences also serve as a barcode to fragment decontamination, which provides high-sensitivity and precise single-cell chromatin accessibility analysis.
Steps
REAGENT SETUP
Prepare METATAC Reagents
ATAC resuspension buffer (ATAC-RSB) 5mL ATAC resuspension buffer (ATAC-RSB)
Mix 50µL 1 M Tris-HCl pH 7.5, 10µL 5 M NaCl, 15µL 1 M MgCl2, and bring the final volume to 5mL with nuclease-free H2O. Store the buffer at -20 °C for up to several months.
| A | B | C |
|---|---|---|
| Reagents | 5 mL | Final conc. |
| 1 M Tris-HCl pH 7.5 | 50 µL | 10 mM |
| 5 M NaCl | 10 µL | 10 mM |
| 1 M MgCl2 | 15 µL | 3 mM |
| Nuclease-free H2O | 4925 µL | |
| Total | 5 mL |
200µL Omni-ATAC lysis buffer
50µL for each reaction, mix 200µL ATAC-RSB with 2µL 10% IGEPAL CA6302µL 10 Tween 202µL 1% digitonin
| A | B | C |
|---|---|---|
| Reagents | 200 µL | Final conc. |
| ATAC RSB | 200 µL | |
| 10% IGEPAL CA630 | 2 µL | 0.1% |
| 10 % Tween 20 | 2 µL | 0.1% |
| 1% Digitonin | 2 µL | 0.01% |
600µL Omni-ATAC wash buffer
150µL for each reaction, mix 600µL ATAC-RSB with 6µL 10% Tween 20. Freshly prepare before use.
| A | B | C |
|---|---|---|
| Reagents | 600 µL | Final conc. |
| ATAC RSB | 600 µL | |
| 20% Tween 20 | 6 µL | 0.1% |
1mL 2 x TD buffer
mix 20µL 1 M TAPS pH 8.5, 10µL 1 M MgCl2, 200µL DMF1mL with nuclease-free H2O. Store the buffer at -20 °C for up to several months.
| A | B | C |
|---|---|---|
| Reagents | 1 mL | Final conc. |
| 1 M TAPS pH 8.5 | 20 µL | 20 mM |
| 1 M MgCl2 | 10 µL | 10 mM |
| DMF | 200 µL | 20% |
| Nuclease-free H2O | 770 µL |
1mL 2 x STOP buffer
mix 80µL 0.5 M EDTA, 10µL 1 M Tris-HCl pH 8.0, 10µL 0.1M spermidine, and bring the final volume to 1mL with nuclease-free H2O. Freshly prepare before use.
| A | B | C |
|---|---|---|
| Reagents | 1 mL | Final conc. |
| 0.5 M EDTA | 80 µL | 40 mM |
| 1 M Tris-HCl pH 8.0 | 10 µL | 10 mM |
| 0.1 M spermidine | 10 µL | 1 mM |
| Total | 1 mL |
50µL Assemble META16 transposome METATAC_Primer_v.1.xlsx
- Anneal META16 transposon
| A | B |
|---|---|
| Oligos | Sequence |
| META16-1 | GGCACCGAAAA |
| META16-2 | CTCGGCGATAAA |
| META16-3 | GGTGGAGCATAA |
| META16-4 | CGAGCGCATTAA |
| META16-5 | AGCCCGGTTATA |
| META16-6 | TCGGCACCAATA |
| META16-7 | GCCTGTGGATTA |
| META16-8 | GCGACCCTTTTA |
| META16-9 | GCATGCGGTAAT |
| META16-10 | GCGTTGCCATAT |
| META16-11 | GGCCGCATTTAT |
| META16-12 | ACCGCCTCTATT |
| META16-13 | CCGTGCCAAAAT |
| META16-14 | TCTCCGGGAATT |
| META16-15 | CCGCGCTTATTT |
| META16-16 | CTGAGCTCGTTTT |
| 19 bp ME | 5'-/phos/-CTGTCTCTTATACACATCT-3' |
| META Tranposon | 5'-[META sequence]-AGATGTGTATAAGAGACAG-3' |
| A | B | C |
|---|---|---|
| Reagents | Per 50 µL | Final conc. |
| 10 x Annealing Buffer | 5 µL | 1x |
| 50 µM META16 Transposon | 5 µL | 5 µM |
| 50 µM 19 bp ME | 5 µL | 5 µM |
| H2O | 35 µL |
Mix thoroughly, then run the annealing program (95 °C, 1 min, gradual cooling, -0.1 °C /3s, 700 cycles to 25 °C, hold at 4 °C)
Recipe for 10x annealing buffer (500 mM NaCl, 100 mM Tris-HCl pH 8.0, 10 mM EDTA)
- Assemble METAT16 transposome
Mix 25µL 5 µM Tn5 transposase and 25µL 5 µM annealed META16 transposon, incubate at 21-24°C for 30 min, protected from light.
Bulk Transposition
Harvest fresh culture in a conical centrifuge tube (15 ml or 50 ml) at room temperature, centrifuge at 500 x g for 5 min at room temperature, then wash twice with 1x PBS pH 7.4, count cell number, stain with Trypan blue, and ensure viability >90%. then aliquot 50, 000 cells to a 200 µL PCR tube.
Pellet 50,000 viable cells at 500 x g at 4°C for 5 min in a swing bucket centrifuge, and remove supernatant carefully without disturbing the pellet.
Add 50 µL ice-cold Omni-ATAC lysis buffer (step 1.2), pipette up and down 10 times, then incubate on ice for 3 min.
Wash out lysis with 150 µL of ice-cold Omni-ATAC wash buffer (step 1.3) and invert the tube 3 times to mix.
Pellet nuclei at 500 x g for 10 min at 4°C in a swing bucket centrifuge.
Then wash one time with 50ul ice-cold Omni-ATAC wash buffer. Pellet nuclei at 500 x g at 4°C for 5min.
Transposition in Bulk
Prepare Transposition mix
| A | B | C | D |
|---|---|---|---|
| Reagents | 3 Rxn | Per Rxn | Final conc. |
| 2 x TD buffer | 37.5 µL | 12.5 µL | |
| META 16 Transposome | 6 µL | 2 µL | 100 nM |
| 1 x PBS | 28.5 µL | 9.5 µL | |
| 1% Digitonin | 0.75 µL | 0.25 µL | 0.01% |
| 10% Tween 20 | 0.75 µL | 0.25 µL | 0. 1% |
Aspirate all supernatant, and avoid disrupting the visible pellet. Then resuspend the cell pellet in 25 µL of transposition mixture by pipetting up and down 10 times, then transfer to a 1.5 mL Lo-bind tube.
Incubate the reaction at 37°C for 30 minutes in a thermomixer with 1000 RPM mixing.
Add 25 µL 2x Stop buffer to stop transposition and incubate on ice for 10 min.
FACS single nuclei
Add 50 µL 0.5% BSA (by dissolving 0.25 g BSA in 50 mL 1x PBS pH 7.4), then add 5 µL 7-AAD to stain nuclei.
FACS sort single 7-AAD positive nuclei to a 96-well PCR plate, containing 1 µL nuclei lysis buffer (10 mM Tris-HCl pH 8.0, 20 mM NaCl, 1 mM EDTA pH 8.0, 15 mM DTT, 0.1% SDS, 60 µg/mL QIAGEN protease).
Amplification
Seal the plate with PCR sealing film (bio-rad, MSB1001), lysis was done by incubating at 65 °C for 15 min.
After lysis, add 1 µL 3% Triton X-100 to quench SDS. Spin down in a plate centrifuge, vortex to mix.
Amplification
Prepare preamplification mix
| A | B | C | D |
|---|---|---|---|
| Reagents | 120 Rxn | Per rxn | Final conc. |
| 2 x High fidelity Q5 master mix | 360 | 3 | 1x |
| 50 µM META16 primer mix | 23.04 | 0.192 | 100 nM each |
| 100 mM MgCl2 | 6 | 0.05 | |
| Nuclease-free H2O | 90.96 | 0.758 | |
| Cell lysate | NA | 2 | NA |
| A | B |
|---|---|
| Oligos | Sequence |
| META 16 Primer | 5'-[META sequence]-AGATGTGTATAAG-3' |
Aliquot 4 µL above preamplification mix to each well, Spin down in a plate centrifuge, vortex to mix.
Preamplification was incubated as
72°C, 5 min
98°C, 30 s
16 Cycles [98°C , 10 s; 62°C, 30 s; 72°C, 1 min]
72°C, 5 min
4°C hold.
Cell barcoding
| A | B |
|---|---|
| Oligos | Sequence |
| META16-ADP1 | 5'-CTTTCCCTACACGACGCTCTTCCGATCT-[CB1]-[META sequence]-AGATGTGTATAAG-3' |
| META16-ADP2 | 5'-GAGTTCAGACGTGTGCTCTTCCGATCT-[CB2]-[META sequence]-AGATGTGTATAAG-3' |
| CB1-1 | GATATG |
| CB1-2 | ATACG |
| CB1-3 | CCGTCTG |
| CB1-4 | TGCG |
| CB1-5 | GAACTCG |
| CB1-6 | ATGTAG |
| CB1-7 | CCCG |
| CB1-8 | TATGT |
| CB1-9 | GAGTAAG |
| CB1-10 | ATCG |
| CB1-11 | CCTAG |
| CB1-12 | TGACCG |
| CB2-1 | ACTCTA |
| CB2-2 | AGAGCAT |
| CB2-3 | GGTATG |
| CB2-4 | TCGATGC |
| CB2-5 | CTACTAG |
| CB2-6 | TATGCA |
| CB2-7 | CACACGA |
| CB2-8 | GTCGAT |
Add 0.45 µL of one of 96 barcode mixes to each well.
Incubate as
98°C, 30 s,
5 cycles [98°C, 10 s, 62°C, 30 s, 72°C, 1 min]
72°C, 5 min
Library preparation
Library Preparation
Prepare Library prep mix
| A | B | C | D |
|---|---|---|---|
| Reagents | 40 Rxn | Per rxn | Final conc. |
| 2x Q5 master mix | 600 | 15 | 1x |
| NEB universal primer(10 µM) | 20 | 2 | 0.67 µM |
| Neb i7 Index primer(10 µM) | 2 | 0.67 µM | |
| 100 mM MgCl2 | 1 | 0.1 | |
| Template | 10.9 | ||
| Total | 30 |
Library preparation was done by incubating as
98°C, 30 s
2 cycle [98°C, 10 s, 68°C, 30 s, 72°C, 1 min]
72°C, 5 min
Purify with ZYMO DCC5, then purify with 1.1 x SPRI beads to remove primer dimers.
Sequencing
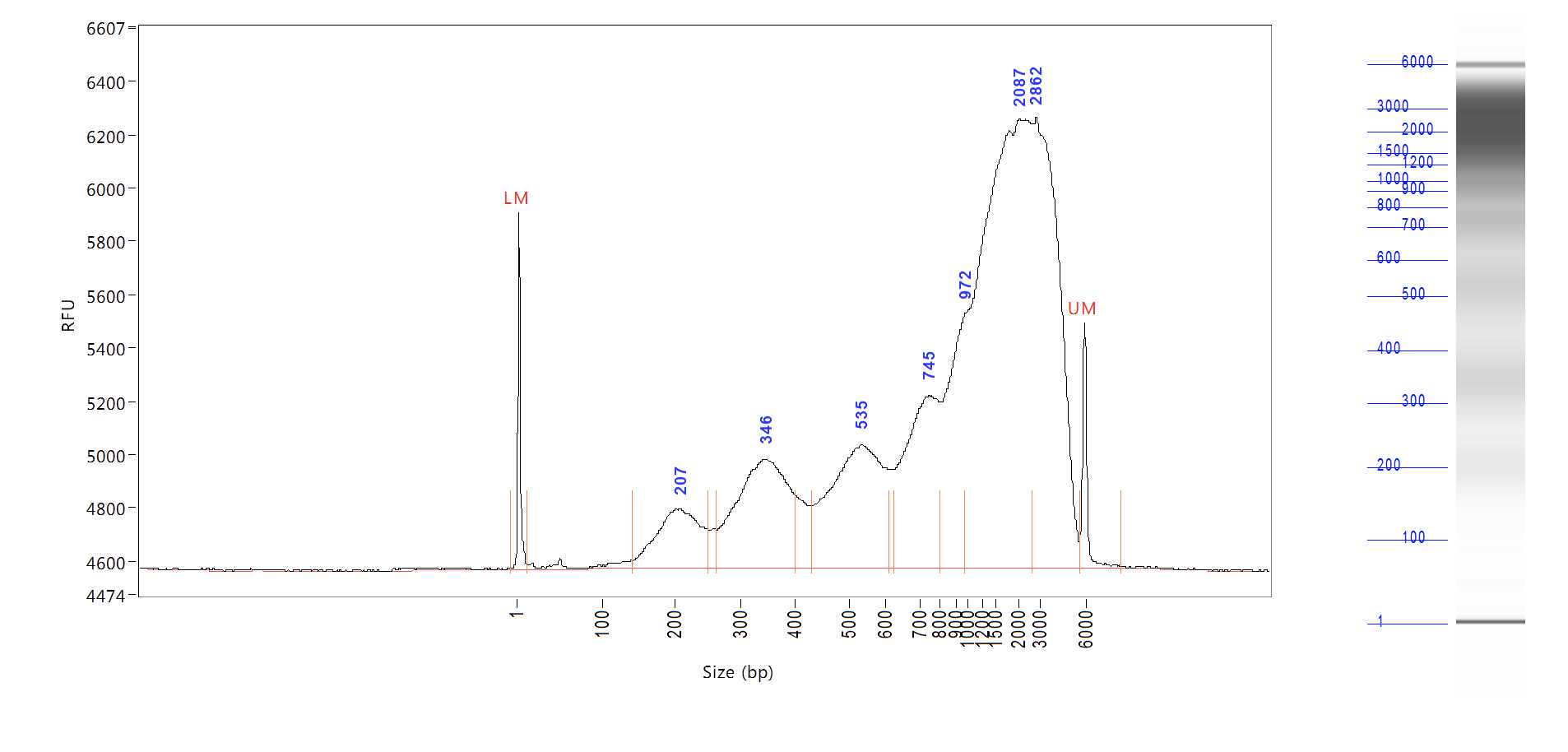
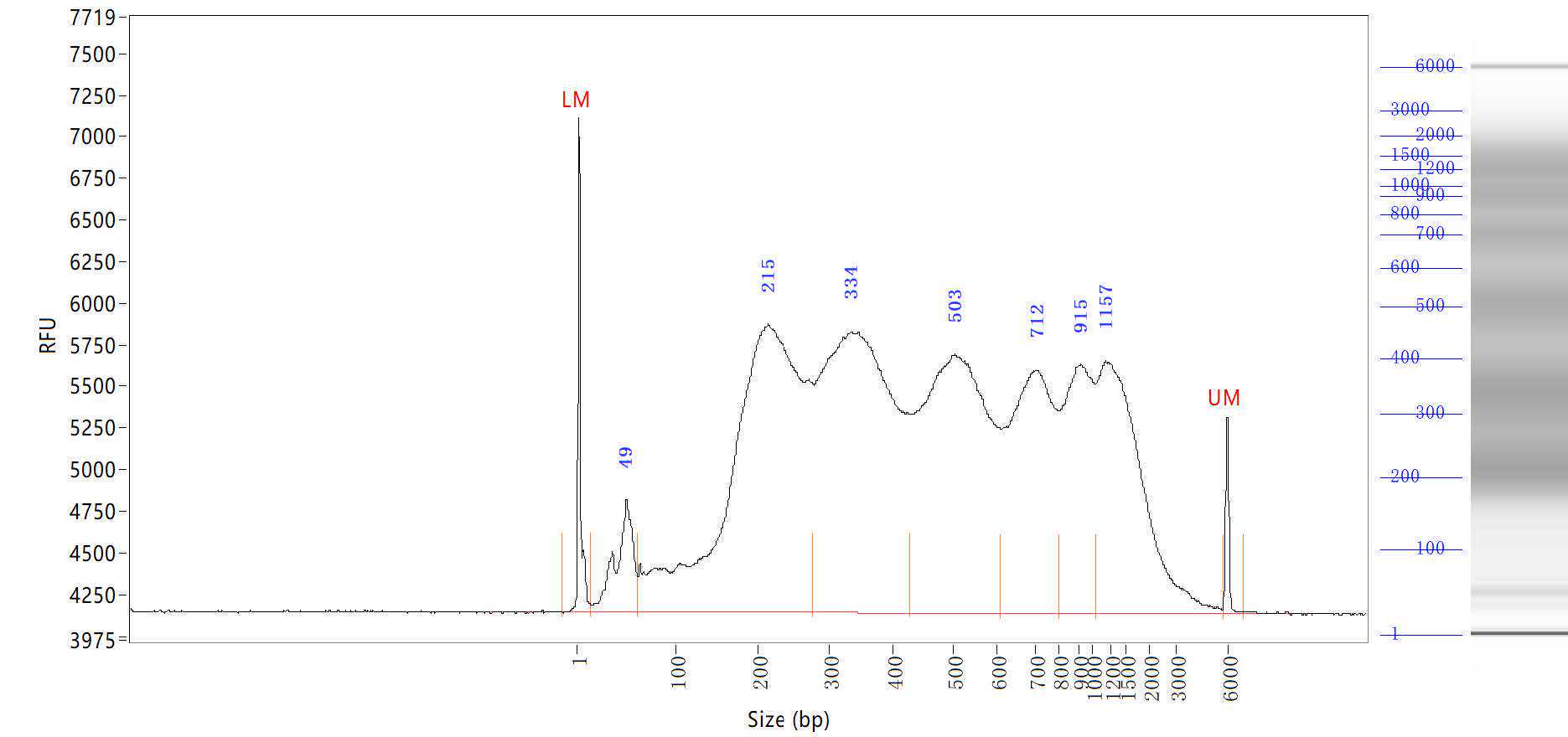
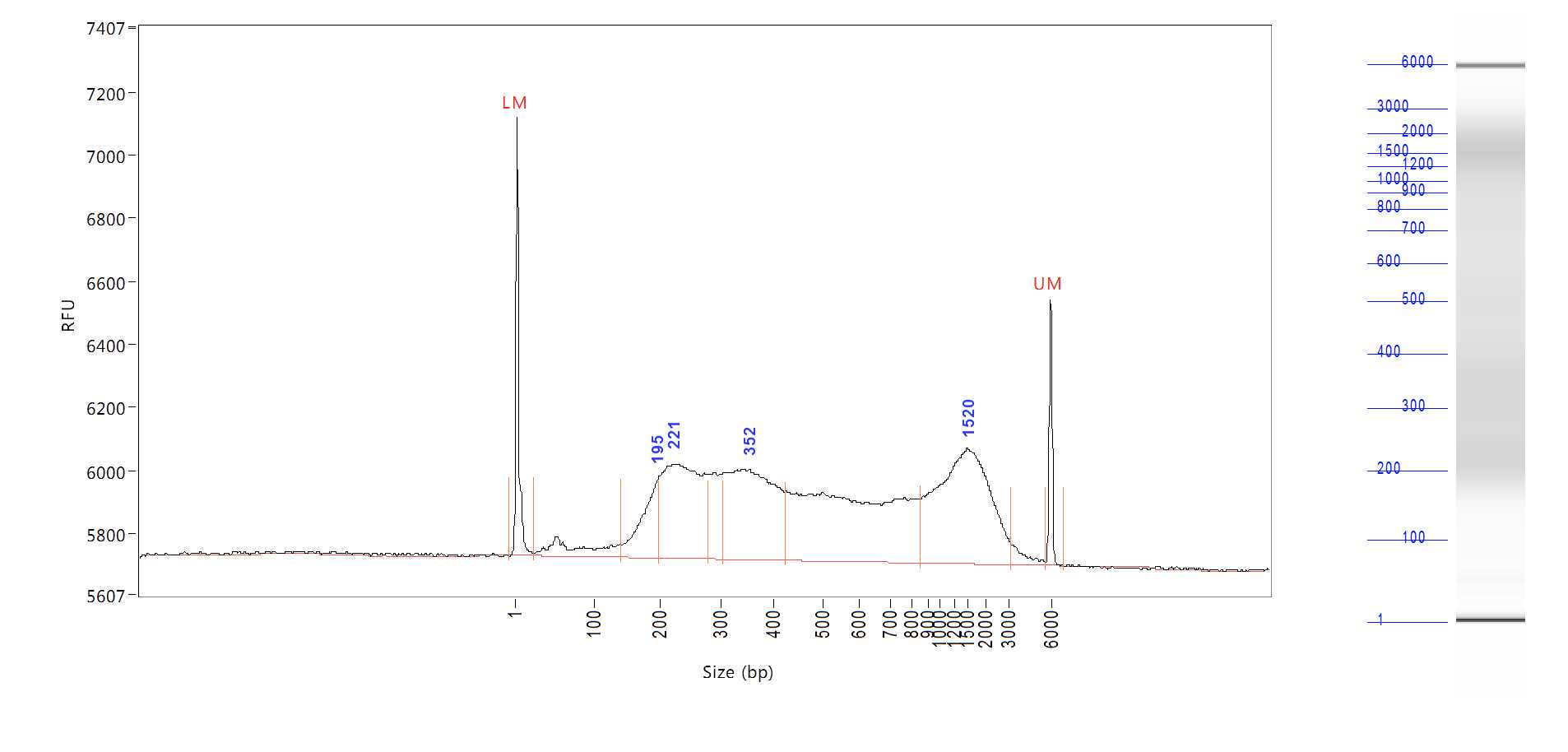
For sequencing, we sequenced our sample on Illumina Hiseq 4000 or NovaSeq sequencer with 9 Gb/plate.
Raw read processing.
Raw Read Preprocessing. For both read 1 and read 2, the first 4 to 7 bases and the following 11 to 13 bases are paired cell barcodes and META sequence, respectively (step 1.6 attachment). We used a custom Python script to parse barcodes and split reads into individual fastq files for each cell, allowing up to one mismatch. Meanwhile, META sequences were annotated to read the name, allowing up to two mismatches. Then we used cutadapt to trim adapter sequences from both ends according to the 19-bp mosaic end (ME) sequence, with parameters -e 0.22 -a CTGTCTCTTATACACATCT and -e 0.22 -g AGATGTGTATAAGAGACAG for both read 1 and read 2. Processed reads were mapped to reference genome with bowtie2 -X 2000 –local –mm –no-discordant –no-mixed. hg38 (GRCh38, v26) reference genome was used for human cells, and mm10 (GRCm38, vM19) reference genome was used for mouse cells. Reads with mapping quality of less than 30 were filtered out from the further analysis. PCR duplicates were identified and removed with a custom script, according to their positions on the genome and META tags. Paired-end reads were converted to fragments with Tn5 insertion centering correction (R1 start +4 and R2 end 5). Finally, for each cell, contaminated fragments from other cells were removed based on the aligned coordinates, META sequences, and read frequency.