Identification, isolation and analysis of human gut-associated lymphoid tissues
Peter B. Jørgensen, Thomas M. Fenton, Urs M. Mörbe, Lene B. Riis, Henrik L. Jakobsen, Ole H. Nielsen, William W. Agace
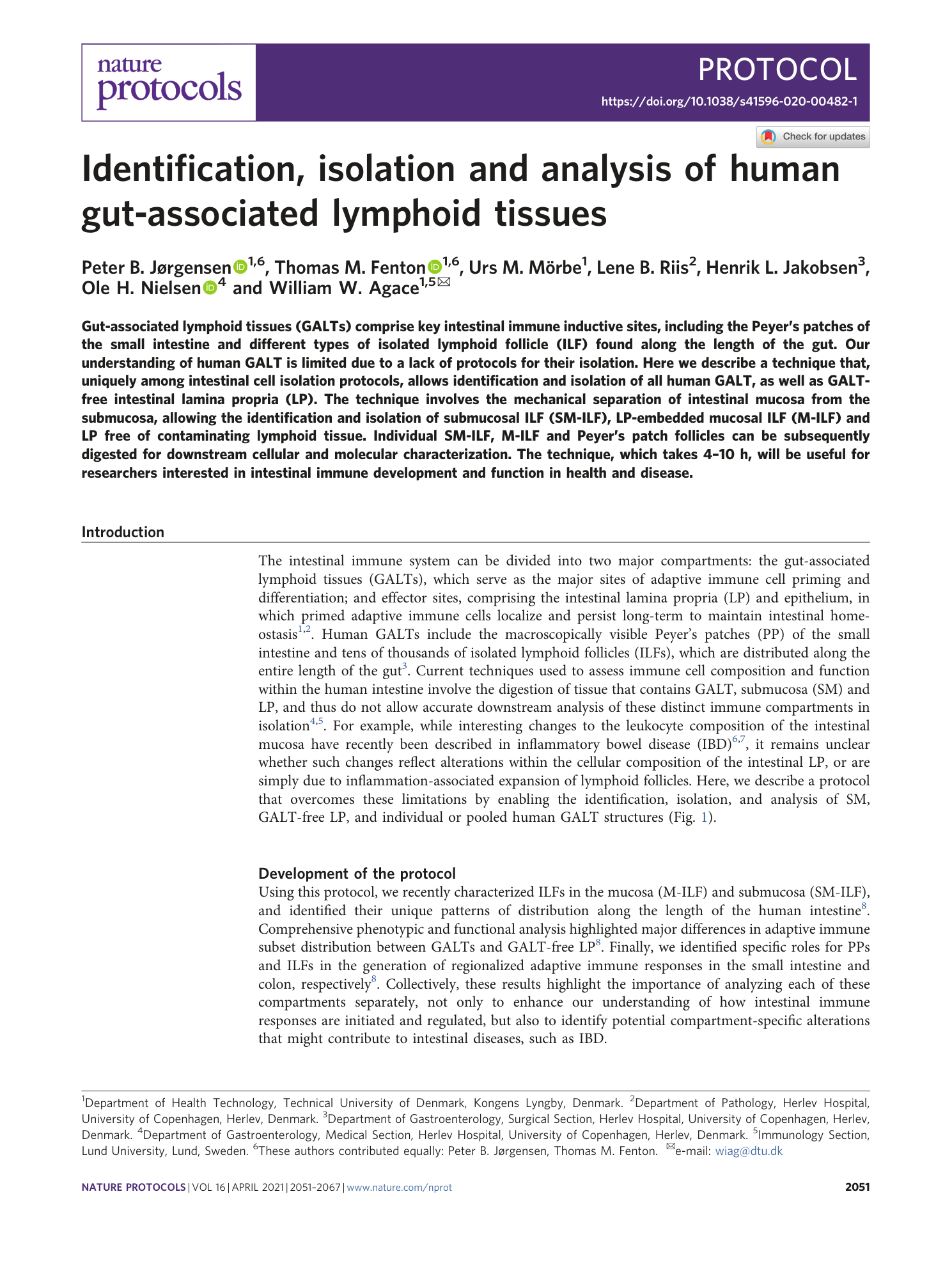
Gut-associated lymphoid tissues
Isolation techniques
Intestinal immunity
Human GALT
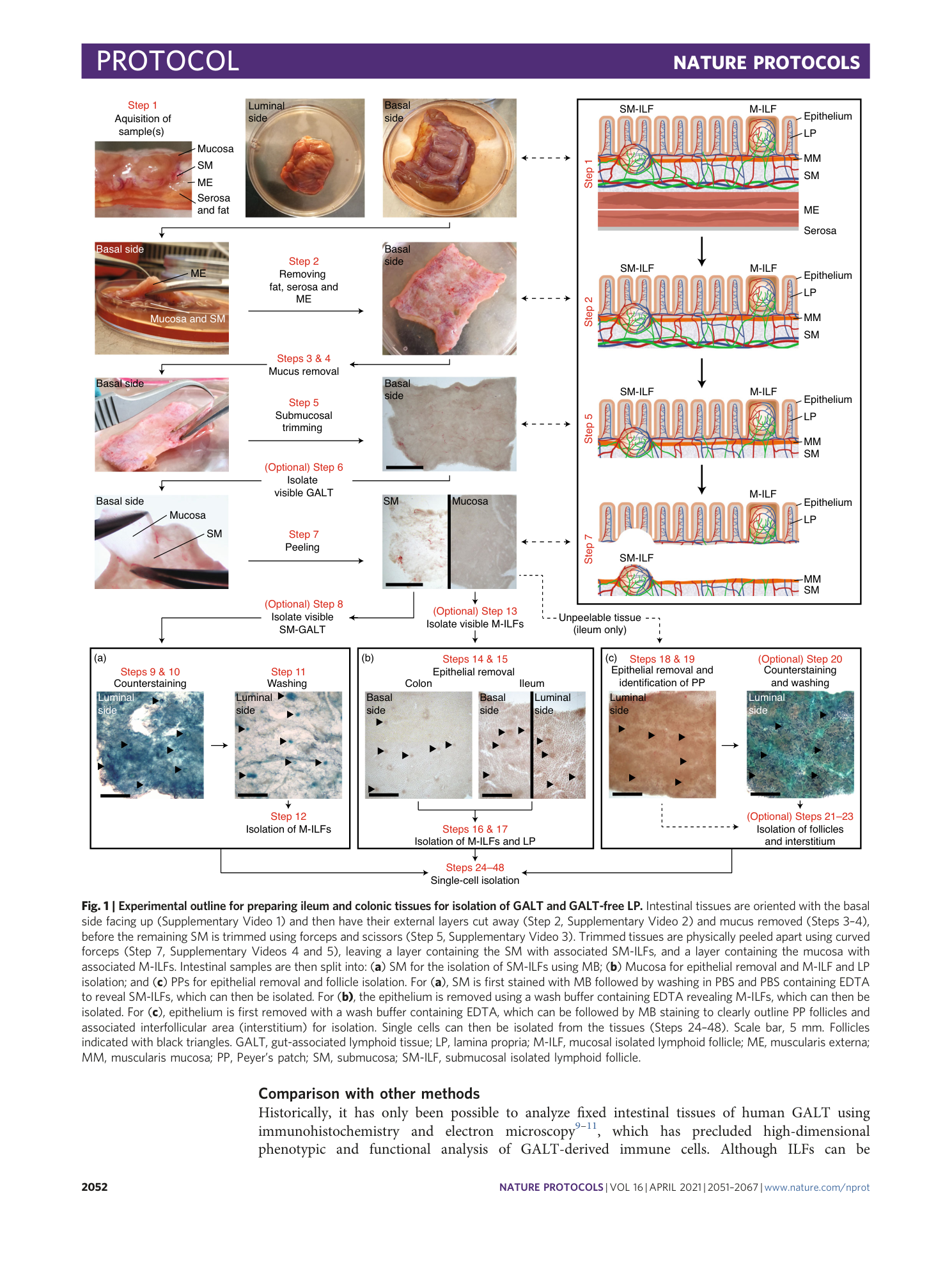
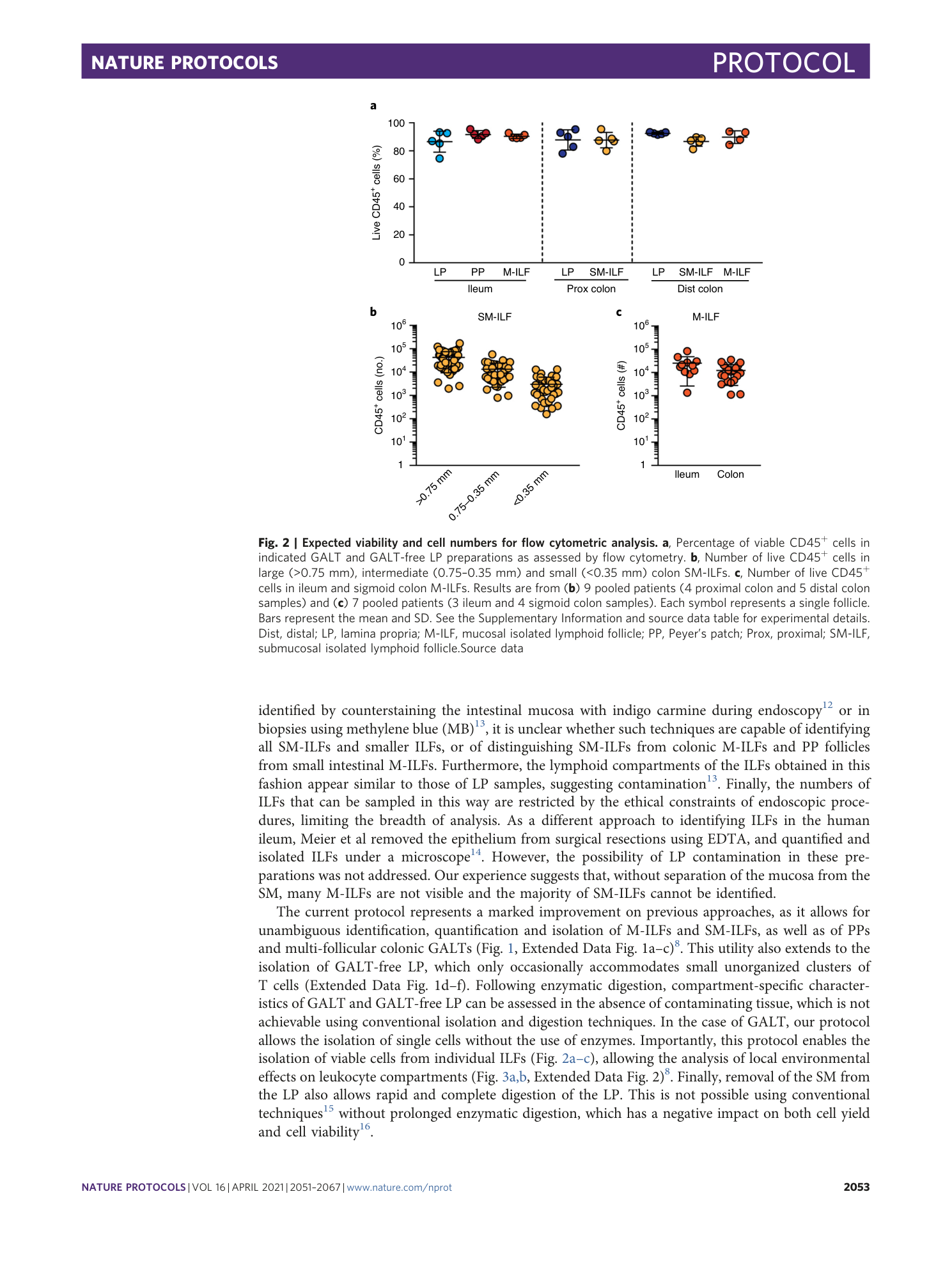
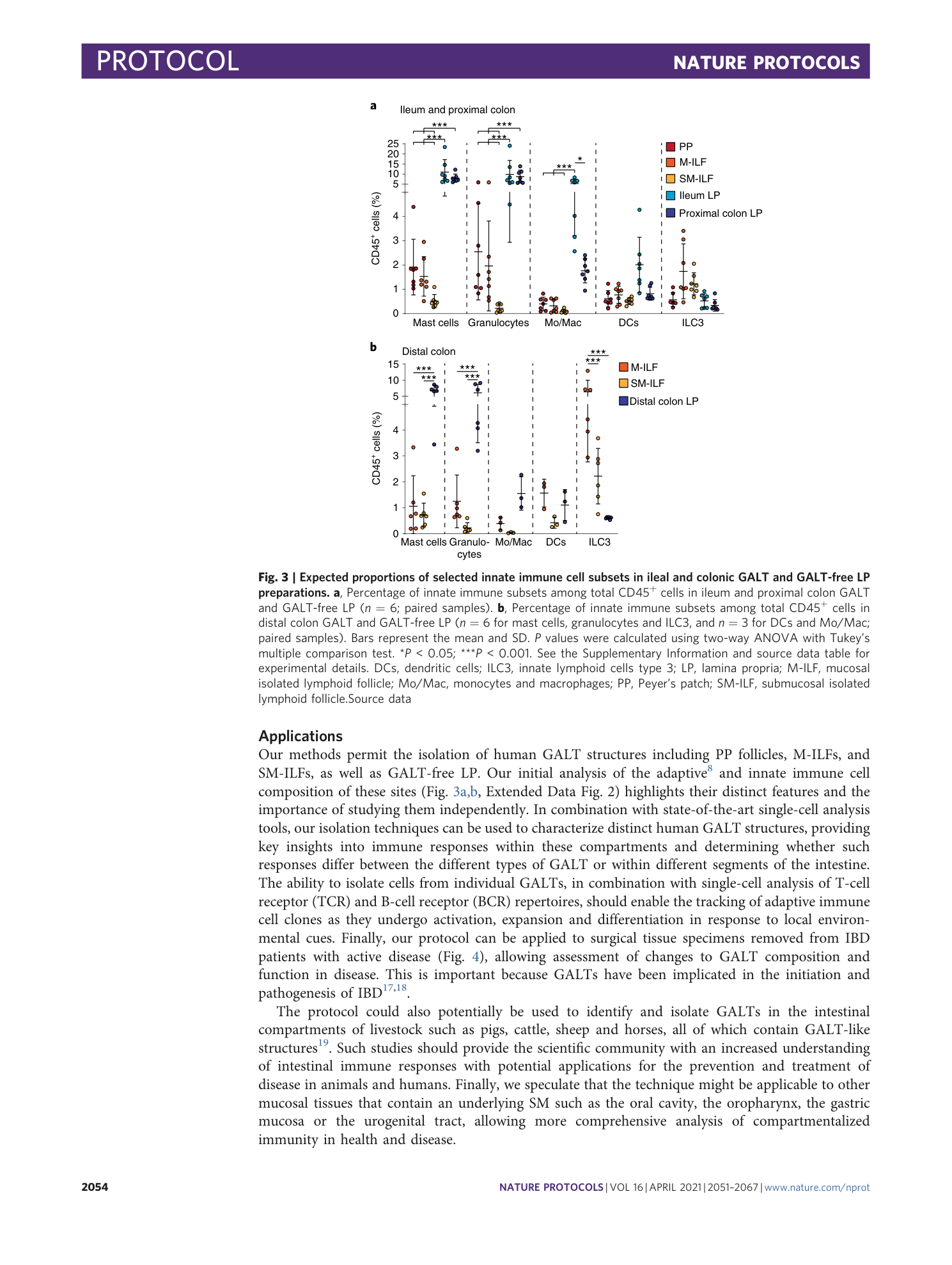
Cellular characterization













Extended
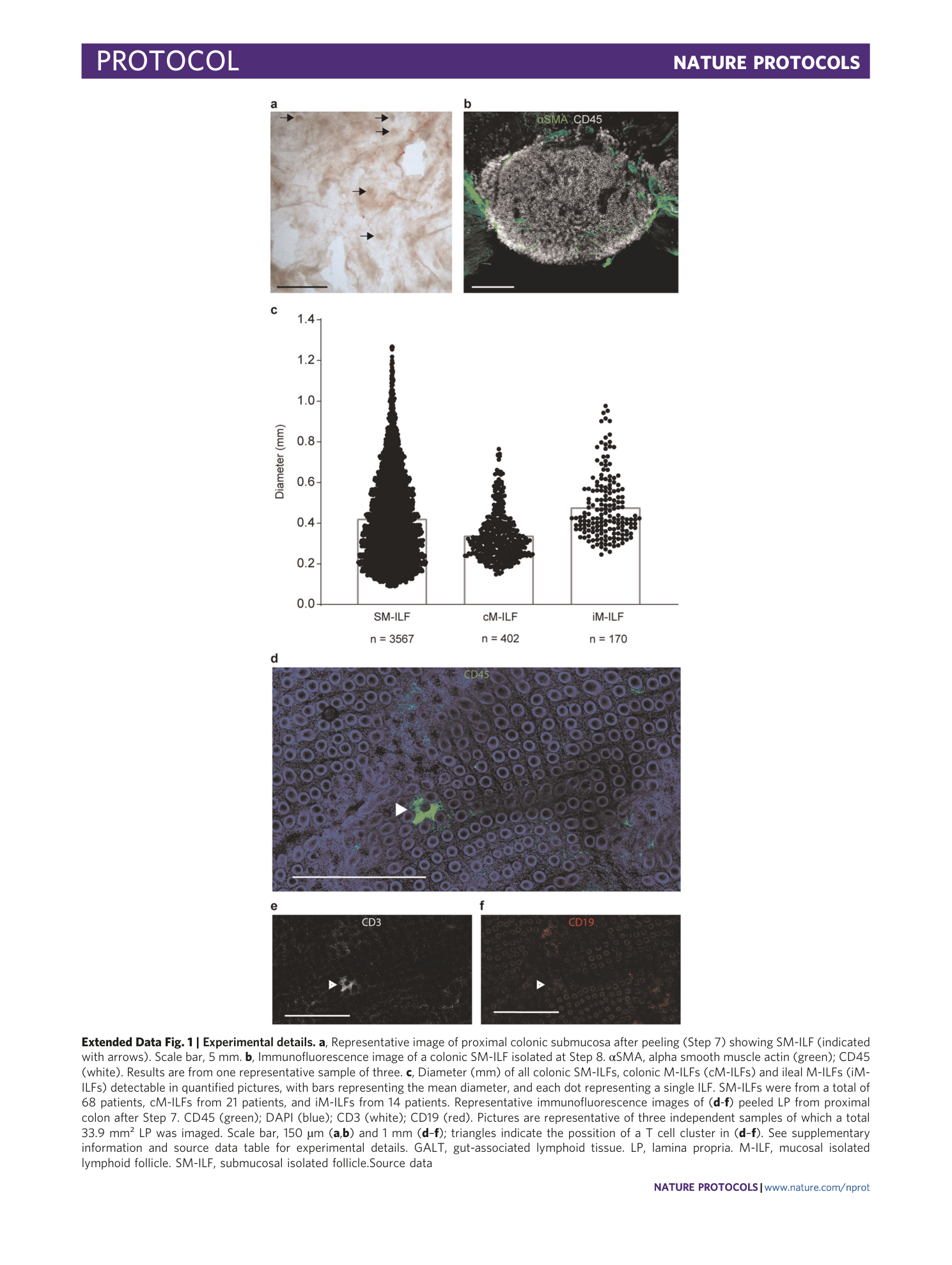
Extended Data Fig. 1 Experimental details.
a , Representative image of proximal colonic submucosa after peeling (Step 7) showing SM-ILF (indicated with arrows). Scale bar, 5 mm. b , Immunofluorescence image of a colonic SM-ILF isolated at Step 8. αSMA, alpha smooth muscle actin (green); CD45 (white). Results are from one representative sample of three. c , Diameter (mm) of all colonic SM-ILFs, colonic M-ILFs (cM-ILFs) and ileal M-ILFs (iM-ILFs) detectable in quantified pictures, with bars representing the mean diameter, and each dot representing a single ILF. SM-ILFs were from a total of 68 patients, cM-ILFs from 21 patients, and iM-ILFs from 14 patients. Representative immunofluorescence images of ( d - f ) peeled LP from proximal colon after Step 7. CD45 (green); DAPI (blue); CD3 (white); CD19 (red). Pictures are representative of three independent samples of which a total 33.9 mm² LP was imaged. Scale bar, 150 µm ( a , b ) and 1 mm ( d – f ); triangles indicate the possition of a T cell cluster in ( d – f ). See supplementary information and source data table for experimental details. GALT, gut-associated lymphoid tissue. LP, lamina propria. M-ILF, mucosal isolated lymphoid follicle. SM-ILF, submucosal isolated follicle.
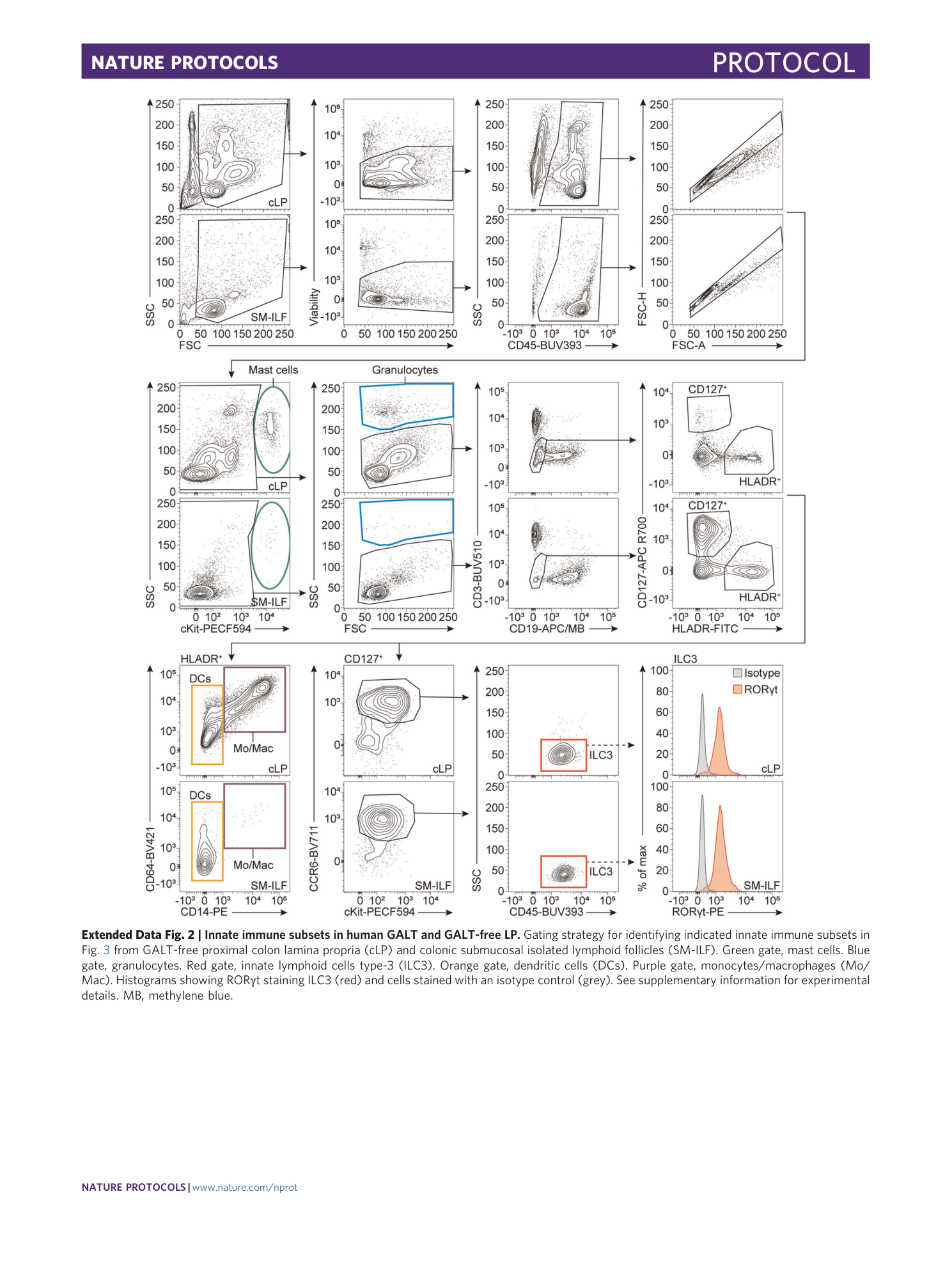
Extended Data Fig. 2 Innate immune subsets in human GALT and GALT-free LP.
Gating strategy for identifying indicated innate immune subsets in Fig. 3 from GALT-free proximal colon lamina propria (cLP) and colonic submucosal isolated lymphoid follicles (SM-ILF). Green gate, mast cells. Blue gate, granulocytes. Red gate, innate lymphoid cells type-3 (ILC3). Orange gate, dendritic cells (DCs). Purple gate, monocytes/macrophages (Mo/Mac). Histograms showing RORγt staining ILC3 (red) and cells stained with an isotype control (grey). See supplementary information for experimental details. MB, methylene blue.
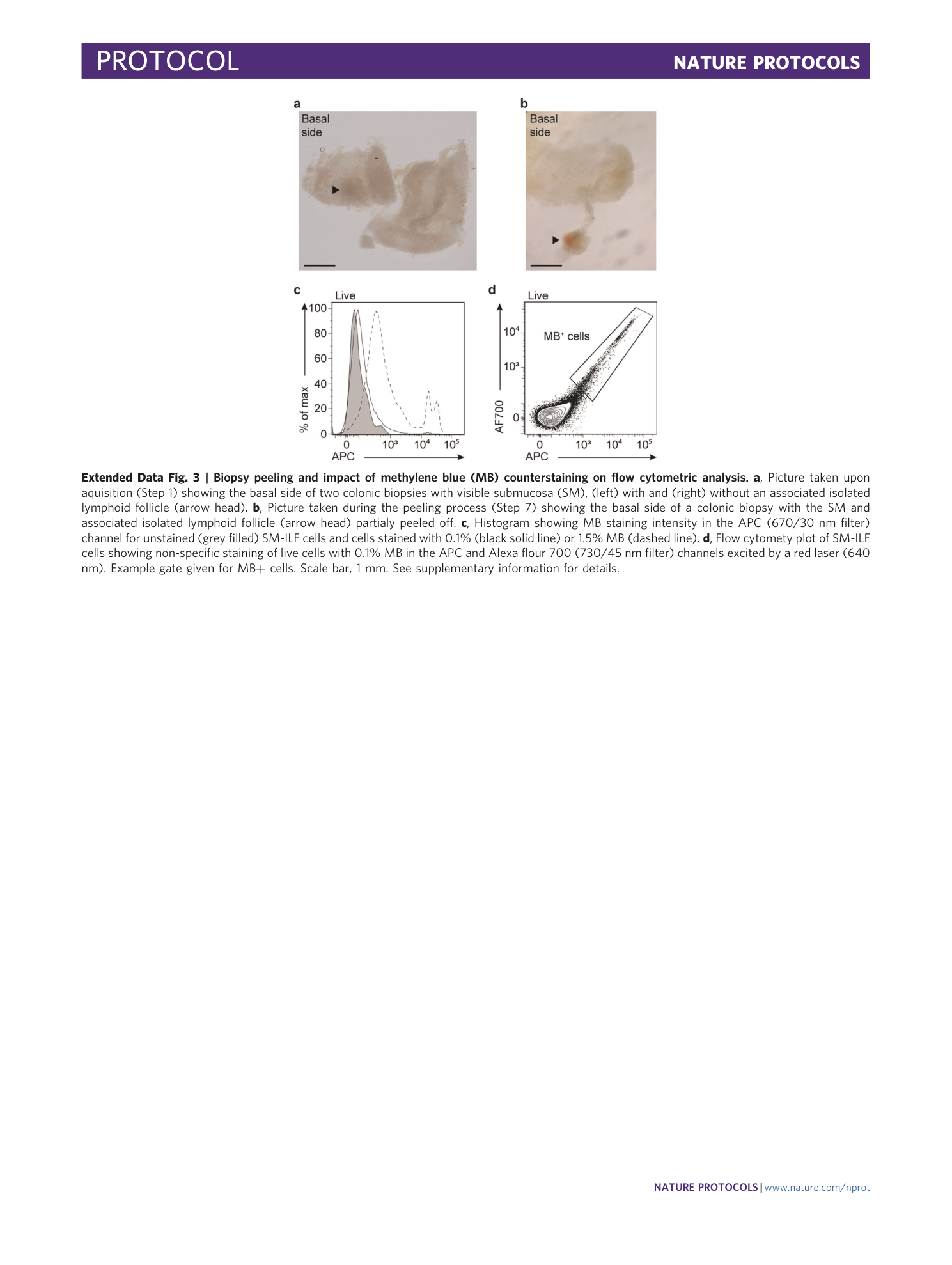
Extended Data Fig. 3 Biopsy peeling and impact of methylene blue (MB) counterstaining on flow cytometric analysis.
a , Picture taken upon aquisition (Step 1) showing the basal side of two colonic biopsies with visible submucosa (SM), (left) with and (right) without an associated isolated lymphoid follicle (arrow head). b , Picture taken during the peeling process (Step 7) showing the basal side of a colonic biopsy with the SM and associated isolated lymphoid follicle (arrow head) partialy peeled off. c , Histogram showing MB staining intensity in the APC (670/30 nm filter) channel for unstained (grey filled) SM-ILF cells and cells stained with 0.1% (black solid line) or 1.5% MB (dashed line). d , Flow cytomety plot of SM-ILF cells showing non-specific staining of live cells with 0.1% MB in the APC and Alexa flour 700 (730/45 nm filter) channels excited by a red laser (640 nm). Example gate given for MB+ cells. Scale bar, 1 mm. See supplementary information for details.
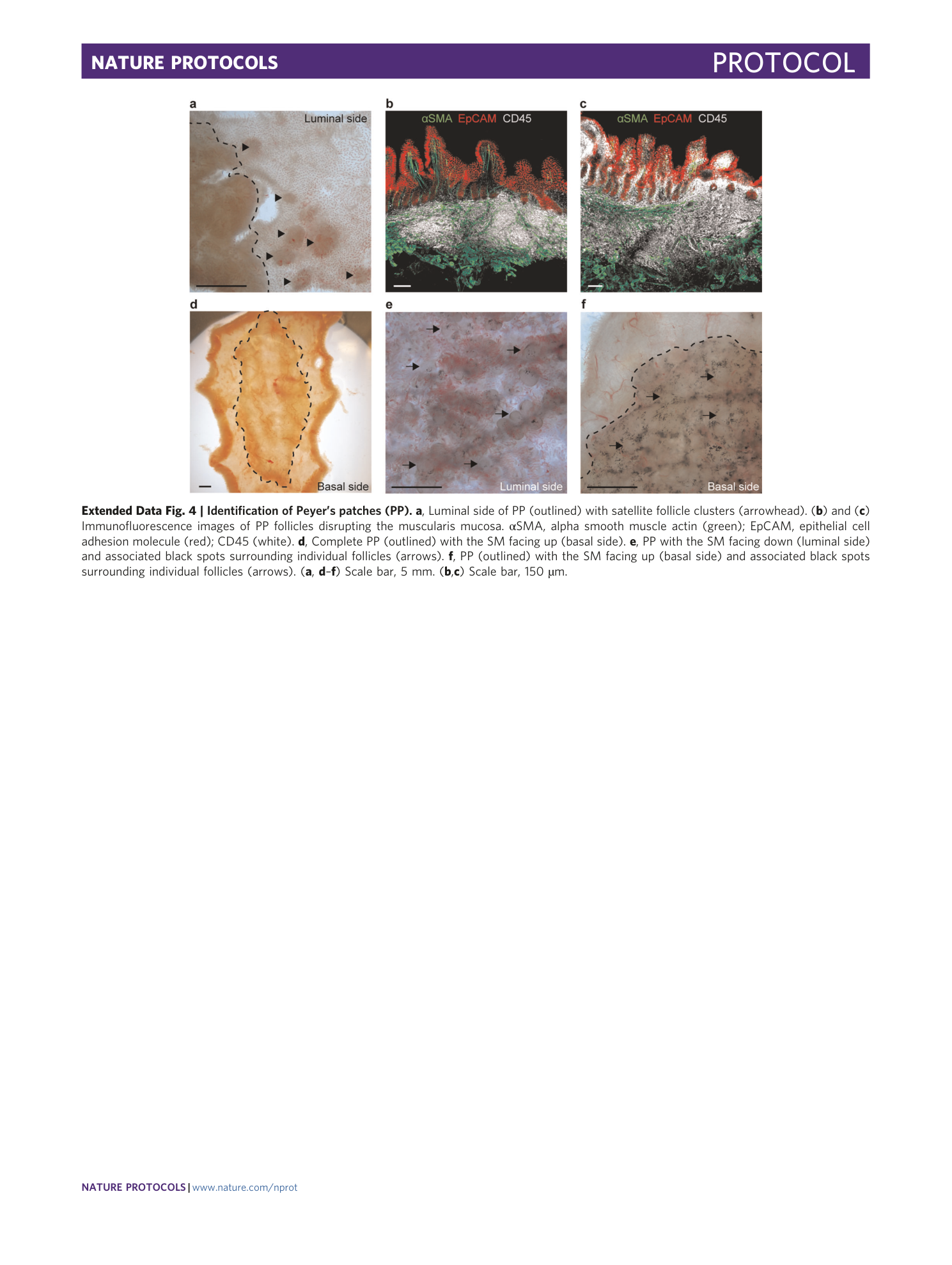
Extended Data Fig. 4 Identification of Peyer’s patches (PP).
a , Luminal side of PP (outlined) with satellite follicle clusters (arrowhead). ( b ) and ( c ) Immunofluorescence images of PP follicles disrupting the muscularis mucosa. αSMA, alpha smooth muscle actin (green); EpCAM, epithelial cell adhesion molecule (red); CD45 (white). d , Complete PP (outlined) with the SM facing up (basal side). e , PP with the SM facing down (luminal side) and associated black spots surrounding individual follicles (arrows). f , PP (outlined) with the SM facing up (basal side) and associated black spots surrounding individual follicles (arrows). ( a , d – f ) Scale bar, 5 mm. ( b , c ) Scale bar, 150 µm.
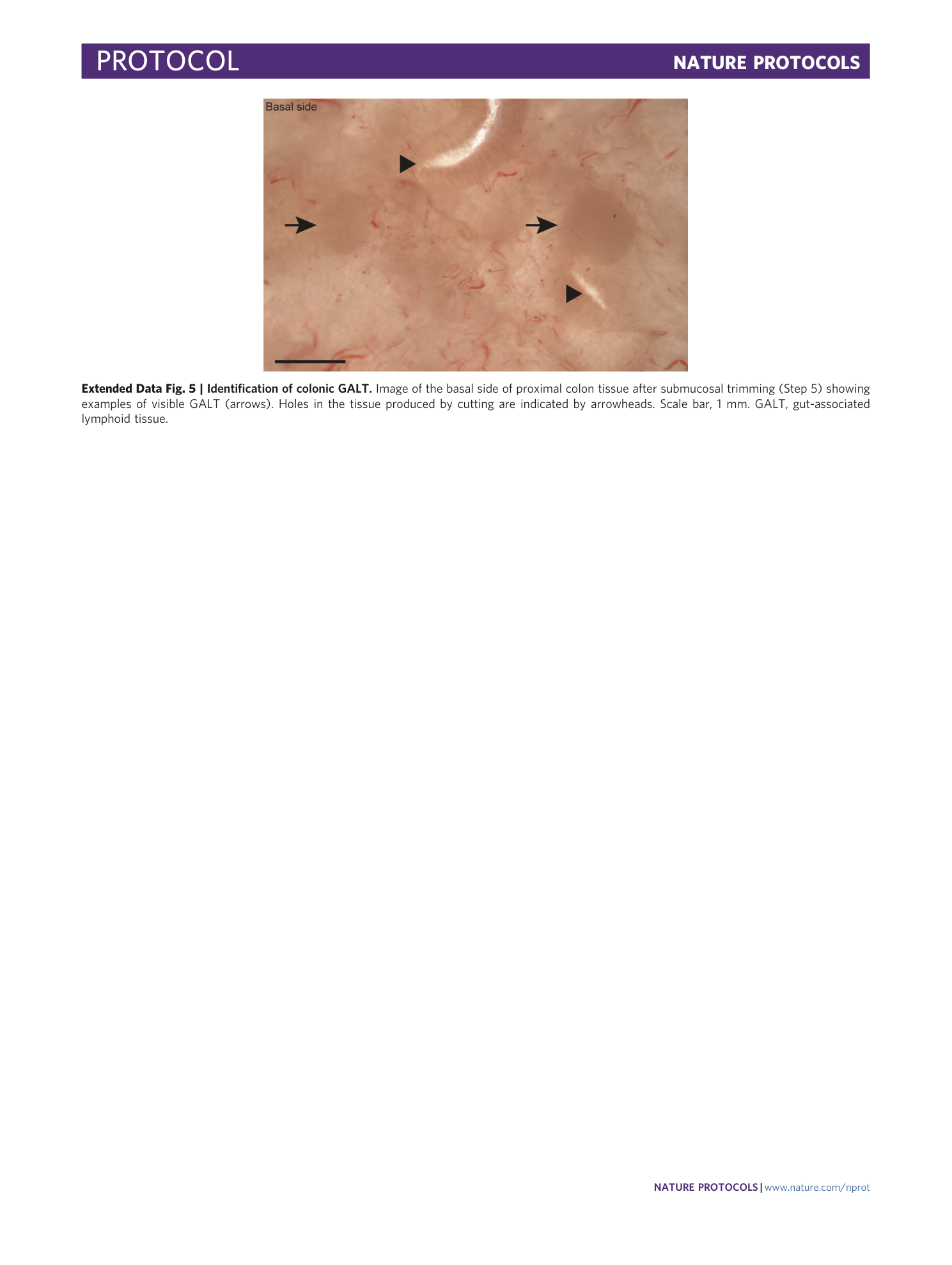
Extended Data Fig. 5 Identification of colonic GALT.
Image of the basal side of proximal colon tissue after submucosal trimming (Step 5) showing examples of visible GALT (arrows). Holes in the tissue produced by cutting are indicated by arrowheads. Scale bar, 1 mm. GALT, gut-associated lymphoid tissue.
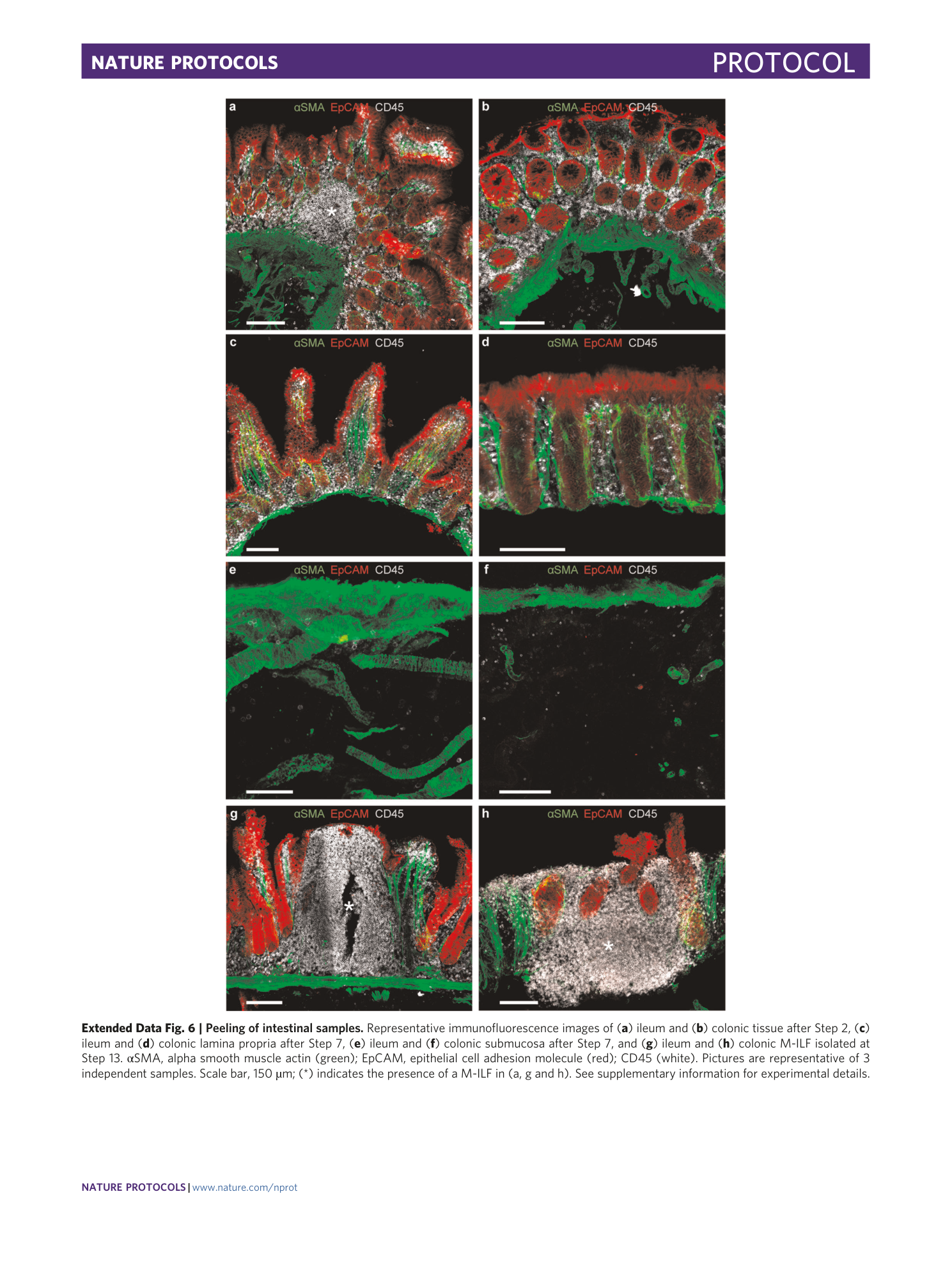
Extended Data Fig. 6 Peeling of intestinal samples.
Representative immunofluorescence images of ( a ) ileum and ( b ) colonic tissue after Step 2, ( c ) ileum and ( d ) colonic lamina propria after Step 7, ( e ) ileum and ( f ) colonic submucosa after Step 7, and ( g ) ileum and ( h ) colonic M-ILF isolated at Step 13. αSMA, alpha smooth muscle actin (green); EpCAM, epithelial cell adhesion molecule (red); CD45 (white). Pictures are representative of 3 independent samples. Scale bar, 150 µm; (*) indicates the presence of a M-ILF in (a, g and h). See supplementary information for experimental details.
Extended Data Fig. 7 Identification of colonic M-ILF and “craters”.
Image of distal colon after peeling (Step 7) showing examples of M-ILF (arrows) and a “crater” (arrowhead) left behind by a peeled submucosal isolated lymphoid follicle. Scale bar, 1 mm. M-ILF, mucosal isolated lymphoid follicle.
Supplementary information
Supplementary Information
Supplementary Table 1, Supplementary Video Legends 1–8 and Supplementary Methods.
Reporting Summary
Supplementary Video 1
Turning over full-thickness colon and ileum wall at Step 1, showing the basal and luminal side of each tissue.
Supplementary Video 2
Cutting off fat, serosa and muscularis externa (Step 2) of colon.
Supplementary Video 3
Trimming submucosa (Step 5) of colon.
Supplementary Video 4
Initiation and peeling (Step 7) of colon.
Supplementary Video 5
Initiation and peeling (Step 7) of ileum.
Supplementary Video 6
Excision of methylene blue stained submucosal isolated lymphoid follicles from the peeled submucosa using a tissue punch (Step 12).
Supplementary Video 7
Identification of a submucosal isolated lymphoid follicle from the basal side, and its association with a mucosal invagination on the luminal side, before the initiation of peeling (Step 6).
Supplementary Video 8
Excision of mucosal isolated lymphoid follicles from peeled sigmoid colon lamina propria using a scalpel (Step 12).

