Homology modeling with trRosetta
Chris Berndsen
Abstract
Protocol for homology modeling using trRosetta written for students in Biochemistry I at James Madison University.
Before start
Have a sequence in FASTA format.
Ex.
protein_seq
MASDTERFFGGYP...
Steps
Setting up modeling
Navigate to the trRosetta homepage.
Enter your email address and give a target name.
Adjust any options that you want and record these changes as a note on this step.
Press submit and wait for the job finish email. Typically takes 2 to 24 hours to get a result.
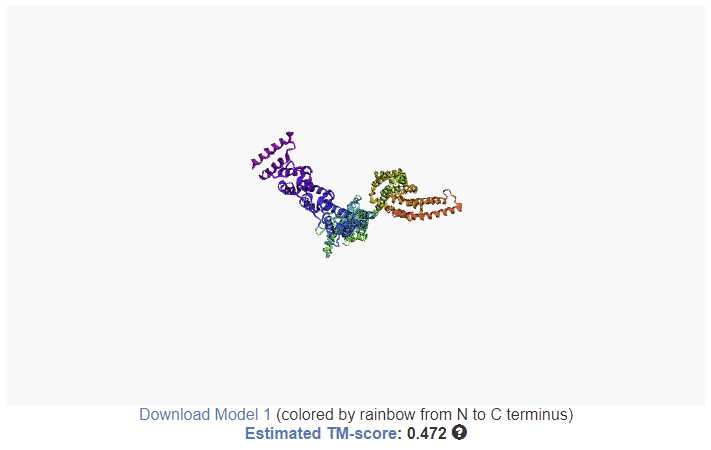
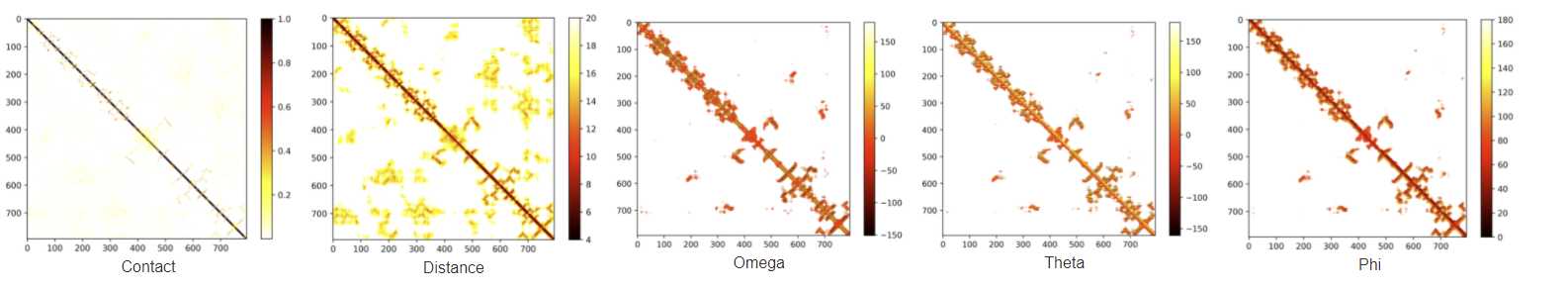
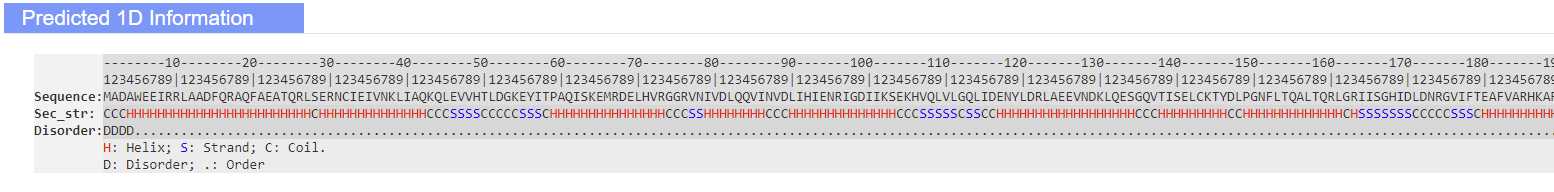
Modeling results
When your receive the job finished email, navigate to the results link.
Saving data
At the top of the window you can download the entire file in .tar.bz2 format. To open this file it must be decompressed.
Alternatively, you can download the relevant models and data individually from the summary table.
Save files as
[date][proteinname][teamname]_trRosetta
Replace [proteiname] with the target protein name, [teamname] with your name or your team's name, [date] with the date.
Ex.: 20210810_UFL1_Berndsen_trRosetta
Indicate WHERE you saved the file as a note on this step.