Home made SM 100bp DNA ladder for agarose gel
Stéphane Mauger
Abstract
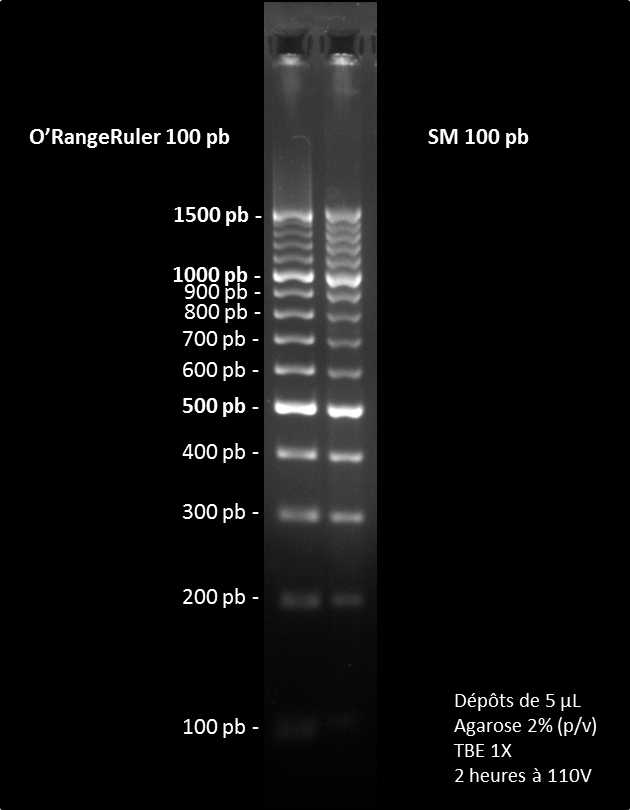
The SM 100pb DNA Ladder is a standard size marker equivalent to the Fisher’s O'RangeRuler® 100 pb DNA Ladder (#SM0623). The SM 100pb Ladder allows to determine the size of double-stranded DNA fragments between 100 bp and 1500 bp and it is composed of 15 double-stranded DNA fragments of 100, 200, 300, 400, 500, 600, 700, 800, 900, 1000, 1100, 1200, 1300, 1400 and 1500 bp. Like the O'RangeRuler 100 bp, the SM 100 bp gives more intense bands at 500, 1000 and 1500 bp.
At the end, the SM 100 pb DNA Ladder is 25 times cheaper than the O'RangeRuler 100 pb.


Steps
Primers and pGEM®-3Zf(+) Vectors preparation
Oligo primers dilution
in 16 1.5mL microtubes, dilute 1:10 each oligo primer at 100µM to final concentration 10µM, see the SM100pb_primers.xlsx file below.
50µL oligo primer (100µM)
450µL nuclease free water
Homogenize and store at 4°C (or at -20°C for a long-term storage).
SM100pb oligo primers sequences (salt free purification):
pGEM®-3Zf(+) Vectors dilution
In 1.5mL microtube, dilute 1:100 pGEM®-3Zf(+) vector at 1µg/µL to final concentration 10ng/µL.
5µL pGEM®-3Zf(+) (1µg/µL)
495µL nuclease free water
Homogenize and store at 4°C (or at -20°C for a long-term storage).
PCR amplification to generate 100pb-1500pb double-stranded DNA fragments
PCR mix preparation
SM100pb is produced using 28 PCRs amplifications in a total of 50µL reaction volume:
1 x 50µL for 600pb, 700pb, 800pb, 900pb, 1100pb, 1200pb, 1300pb and 1400pb fragments
2 x 50µL for 200pb, 300pb and 400pb fragments
3 x 50µL for 1000pb and 1500pb fragments
4 x 50µL for 100pb and 500pb fragments
Defreeze and vortex all reagents, except enzymes (stored at -20°C), for approximately0h 0m 5s
Spin down all reagents for approximately 0h 0m 5s and place On ice .
In 1.5µL microtube, prepare the PCR mix according to the following table :
| A | B | C | D | E |
|---|---|---|---|---|
| Initial concentration | Final concentration | n=1 | n=28 | |
| SMF forward primer | 10µM | 400mM | 2µL | 56µL |
| Green GoTaq buffer | 5X | 1X | 10µL | 280µL |
| MgCl2 | 25mM | 1mM | 2µL | 56µL |
| pGEM®-3Zf(+) vecto | 10ng/µL | 20ng | 2µL | 56µL |
| dNTP mix | 2.5mM | 150µM each | 3µL | 84µL |
| GoTaq polymerase | 5 u/µL | 1.75U | 0.35µL | 9.8µL |
| nuclease free water | 28.65µL | 802.2µL | ||
| TOTAL | 48µL | 1344µL |
PCR mix composition
Reverse primers and mix combinaison
Defreeze and vortex all the 15 reverse oligo primers at 10µM
In a 96-well plate PCR, transfer 2µL of each reverse oligo primers (10µM) according to the following map :
| A | B | C | D | E | F | G | H |
|---|---|---|---|---|---|---|---|
| 600R | 200R | 1000R | 500R | ||||
| 700R | 200R | 1500R | 500R | ||||
| 800R | 300R | 1500R | 500R | ||||
| 900R | 300R | 1500R | 500R | ||||
| 1100R | 400R | 100R | |||||
| 1200R | 400R | 100R | |||||
| 1300R | 1000R | 100R | |||||
| 1400R | 1000R | 100R |
Map of the PCR plate
Vortex and spin down the PCR mix tube, transfer 48µL in each of the 28 wells.
Seal the PCR plate.
In thermocycler, run PCR amplification with cycles follows:
| A | B | C | D |
|---|---|---|---|
| Cycles step | Temperature | Time | Cycles |
| Initial denaturation | 94°C | 5 min | 1 |
| Denaturation | 94°C | 30 sec | 40 |
| Annealing | 60°C | 30 sec | 40 |
| Extension | 72°C | 30 sec | 40 |
| Final extension | 72°C | 60 min | 1 |
| Hold | 4°C |
PCR program
After PCR, pool and dilute 1:2 the PCR amplification
In a 5mL tube:
1400mL of PCR amplification (28 x 50µL)
420µL 5X green GoTag buffer
980µL nuclease free water
Homogenize and store at 4°C (or at -20°C for a long-term storage).
Load 5µL to 10µL per line in a agarose gel.