High-throughput workflow for the genotypic characterization of transposon library variants
Ana Mariya Anhel, Lorea Alejaldre, Ángel Goñi-Moreno
High-throughput
Transposon library
marC9
Tn5
SEVA
OT-2
Opentrons
genotypic characterization
genotyping
bacterial genome
96-well
Abstract
This is a workflow for the genotypic characterization of transposon library variants. It has been developed using an open-source Opentrons OT-2 robot, BLASTN for genomic annotations and modular sub-protocols (e.g., PCR sample preparation, OT-2 volume transfer, OT-2 counter selection, etc) that can be used for other tasks, thus providing a general-purpose pipeline.
All steps follow a 96-well plate format for high-throughput analysis. The protocol is described for the characterization of transposon library variants generated with SEVA-Sib pBAMD1-x and pBLAM1-x plasmid sets that follow Standard European Vector Architecture (SEVA, https://seva-plasmids.com) and can be amplified with the standard PS1-PS6 primers. After the description of the protocol we present the results of an example generated at our laboratory (https://biocomputationlab.com) using the soil bacterium Pseudomonas putida KT2440 as acceptor strain.
Steps
Colony picking in selective media
Dispense 100µL of selective media (M9-citrate for P. putida or Luria-Bertani plus 20 ng/µL nalidixic acid for DH5 α E. coli ) plus transposon cassette antiobiotic in a 96-well plate
Pick individual colonies into a 96-well plate with selective media
Tip: Keep tips inside of wells to keep track
Cover with a sterile breathable membrane
Grow at 30°C ( P. putida ) or 37°C ( E. coli ) / 500rpm
Counter-selection and glycerol stocks pre-cultures
Measure OD600nm of overnight culture grown in selective media from plus transposon cassette antibiotic in a plate reader
Inoculation of counter-selection plate in selective media:
- Dispense
100µLof selective media (M9-citrate for P. putida or Luria-Bertani plus 20 ng/µL nalidixic acid for DH5α E. coli ) plus ampicillin (backbone antibiotic) to select against spurious integration events. - Transfer
5µLof overnight culture from to counter-selection (ampicillin) plate - Cover with a sterile breathable membrane
Note
For steps 6 and 7, if two or more 96-well plates are used as input it is advised to use the OT-2 protocol below to minimize human error. Dispensed volume and culture volume inoculated should be that described in these steps. Note that steps 6 and 7 could be completed together in a single run depending on the number of initial plates.
Inoculation of precultures for glycerol stock in rich media:
- Dispense
100µLof Luria-Bertani media plus transposon cassette antiobiotic in a 96-well plate - Transfer
5µLof overnight culture from to counter-selection (ampicillin) plate - Cover with a sterile breathable membrane
Grow counter-selection and glycerol stock pre-culture plates at 30°C ( P. putida ) or 37°C ( E. coli ) / 500rpm
Measure OD600nm of overnight culture grown in selective media plus ampicillin in a plate reader
Colony selection in OT-2 liquid handler robot
Selection of colonies to store as glycerol stocks and do further PCR reactions by running the following OT-2 protocol with its corresponding template.csv
Cover glycerol stock plates with a storage membrane and store at -80ºC
If not proceeding to the next step right away: Store "PCR plate" at 4ºC for a few days or cover with an storage membrane and store at -20ºC for longer term
Master 96-well plate for PCR steps
Transfer 50µL of selected colonies from one or more libraries to a 96-well plate with the following OT-2 protocol:
OT-2 Protocol to transfer volume from several plates to a single plate
Control PCRs
Spurious integration control with SEVA primers pairs PS3/PS4 and PS5/PS6
OT-2 PCR sample preparation protocol
Optional: Cargo integration control with primers PSMCS and either ME-O-Km-R/ME-O-Sm-R or ME-O-Gm-R (depending on transposon cassette antibiotic )
Arbitrary PCRs
Arbitrary PCR#1 using primer pairs ARB6 and ME-O-Km-Ext-F/ME-O-Sm-Ext-F or ME-O-Gm-Ext-F depending on transposon antibiotic cassette
OT-2 PCR sample preparation protocol
Seal 96-well plate, place it in thermocycler and run the following PCR program:
| A | B | C |
|---|---|---|
| 98ºC | 5 min | |
| 98ºC | 10 s | x6 cycles |
| 30ºC | 30 s | |
| 72ºC | 1 min 30 s | |
| 98ºC | 10 s | x30 cycles |
| 45ºC | 30 s | |
| 72ºC | 1 min 30 s | |
| 72 ºC | 5 min | |
| 4ºC | hold |
Select 8-12 Arbitrary PCR#1 reactions from the 96-well plate and run them on a 1% agarose gel to verify amplification.
Arbitrary PCR#2 using primers pairs ARB2 and ME-O-Km-Int-F/ME-O-Sm-Int-F or ME-O-Gm-Int-F depending on transposon antibiotic cassette
OT-2 PCR sample preparation protocol
Seal 96-well plate, place it in thermocycler and run the following PCR program:
| A | B | C |
|---|---|---|
| 98ºC | 30 s | |
| 98ªC | 10 s | x30 cycles |
| 52ºC | 30 s | |
| 72ºC | 1 min 30 s | |
| 72ºC | 5 min | |
| 4ºC | hold |
Select 8-12 Arbitrary PCR#2 reactions from the 96-well plate and run them on a 1% agarose gel to verify amplification
Sequencing and annotation
Prepare a PCR plate to send to sequencing by mixing 10µL of unpurified Arbitrary PCR#2 reaction and 10µLof 10 µM sequencing primer (ME-O-Km-Ext-F/ME-O-Sm-Ext-F or ME-O-Gm-Ext-F depending on the transposon antibiotic cassette)
Annotate sequencing results by running the following protocol:
Bacterial genome annotation script using BLASTN
Example
In this section we show a specific example on how to run the workflow with the OT-2 liquid handler using a starting point three 96-well plates with P. putida KT2440 colonies picked from matings using cargos with either a kanamycin (pBLAM1-2), streptomycin (pBLAM1-4) or gentamicin (pBLAM1-6) resistance gene.
Counter-selection and glycerol stocks pre-cultures s using the following OT-2 protocol in in two runs
The following template. csv will dispense M9-citrate media containing ampicillin to three plates. Subsequently, each plate will be inoculated from each of the three starting plates.
The following template. csv will dispense LB media with either gentamicin, kanamycin or streptomycin to each plates. Subsequently, each plate will be inoculated from each of the three starting plates.
Colony selection in OT-2 liquid handler robot.
Each LB plus cargo antibiotic plate will be run separately with the script in . For each library there is an input .csv file for the absorbance at 600nm in M9-citrate with either kanamycin, gentamicin or streptomycin and another .csv file for the absorbance at 600nm in M9-citrate ampicillin. The LB plate from step 25.2 will be used to inoculate the two glycerol stock plates and PCR plate.
pBLAM1-2: .csv files and template .csv to run script
Variables-ColonieScreening-OT.csv
220920_M9Cit_ant_LibpBLAM12.csv
220921_M9Cit_amp_LibpBLAM12.csv
Output from the run (map with identifiers)
pBLAM1-4: .csv files and template .csv to run script
Variables-ColonieScreening-OT.csv
220920_M9Cit_ant_LibpBLAM14.csv
220921_M9Cit_amp_LibpBLAM14.csv
Output from the run (map with identifiers)
pBLAM1-6: .csv files and template .csv to run script
Variables-ColonieScreening-OT.csv
220920_M9Cit_ant_LibpBLAM16.csv
220921_M9Cit_amp_LibpBLAM16.csv
Output from the run (map with identifiers)
Master 96-well plate for PCR steps to combine cultures from different transposon libraries in a single 96-well plate
The following template.csv is to be run with the OT-2 script in
Variables-SamplesMerging-OT.csv
Output from the run (map with identifiers)
Control PCRs to account for spurious integrations and the correct integration of the cargo
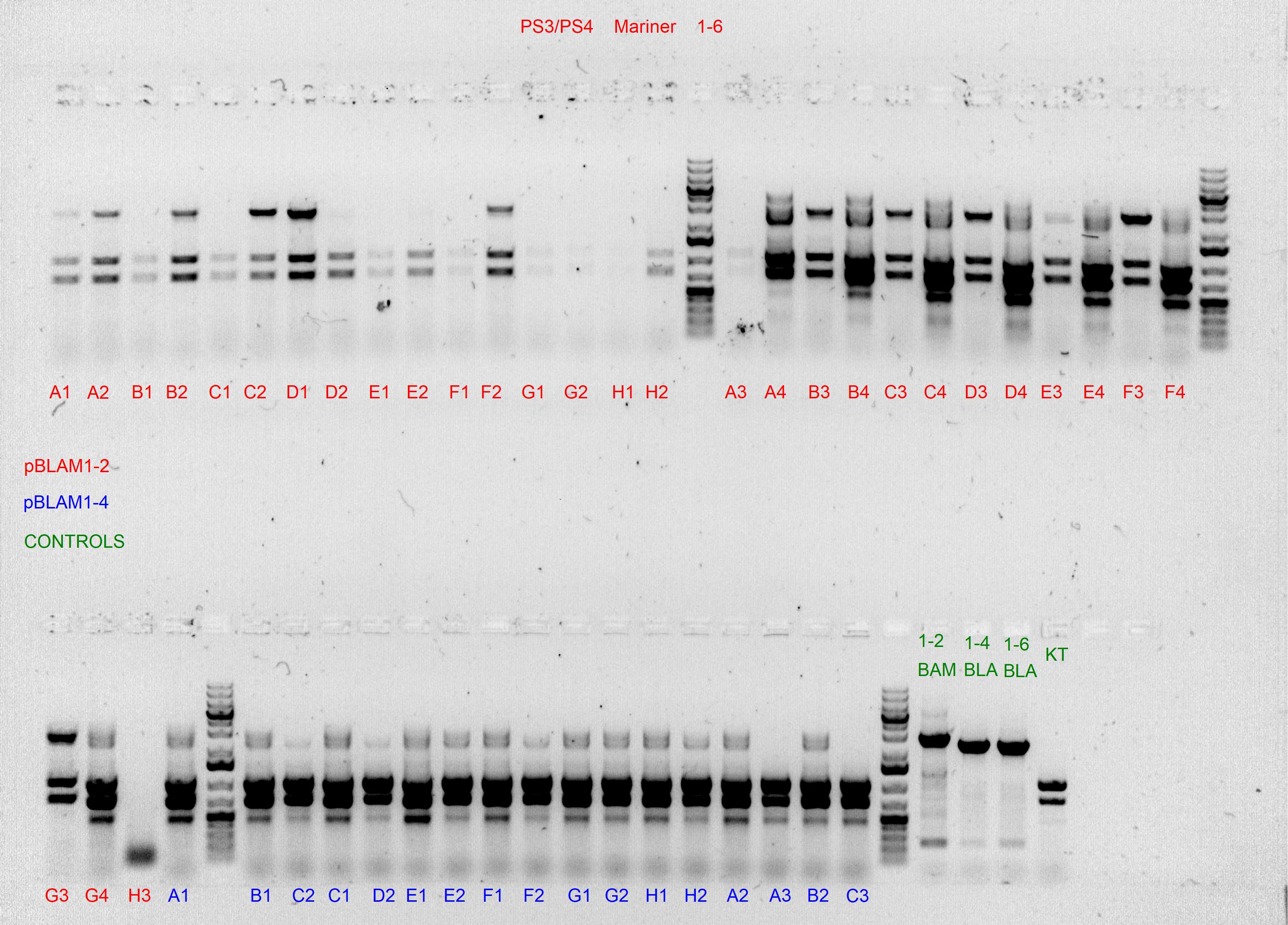
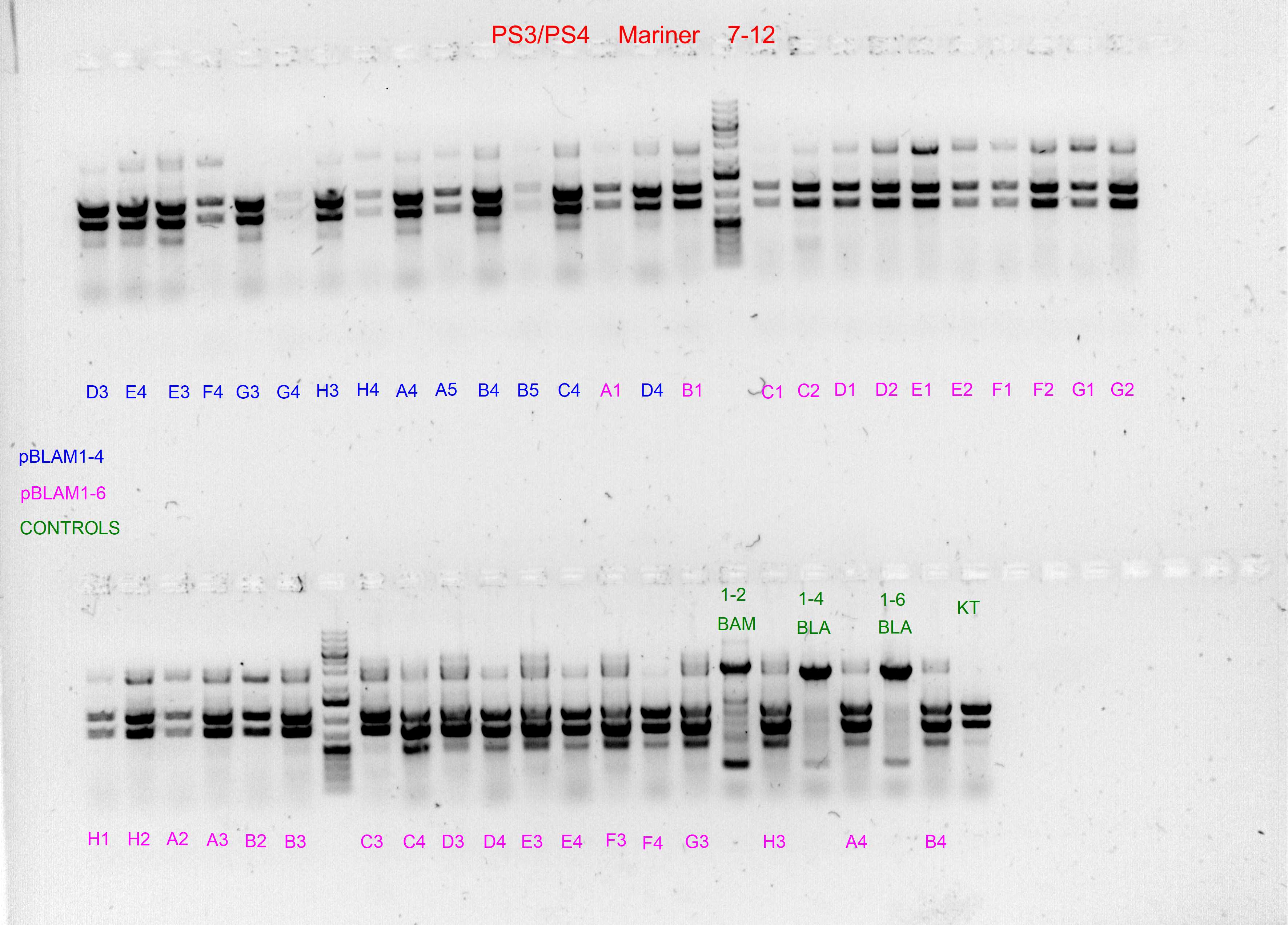
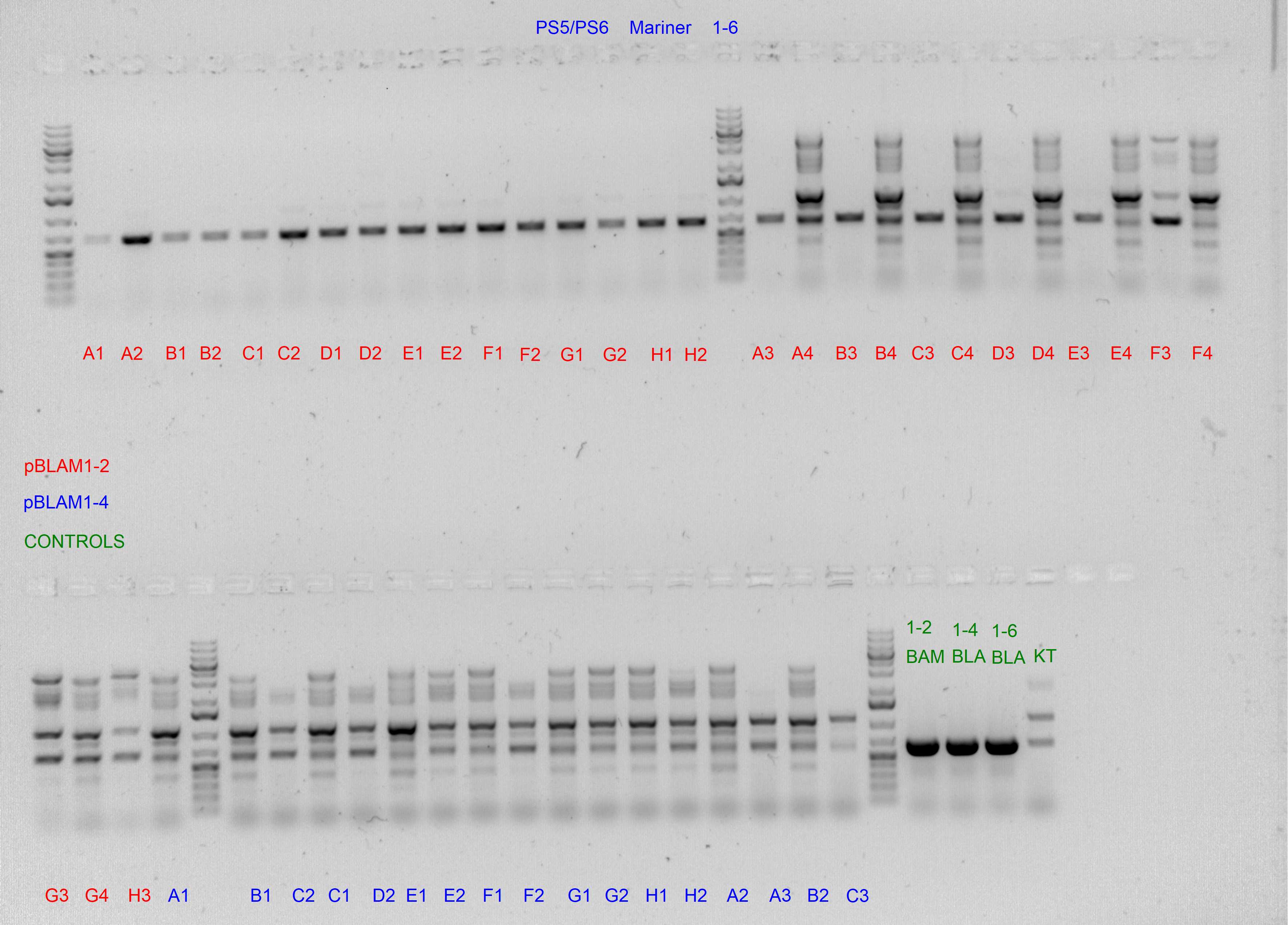
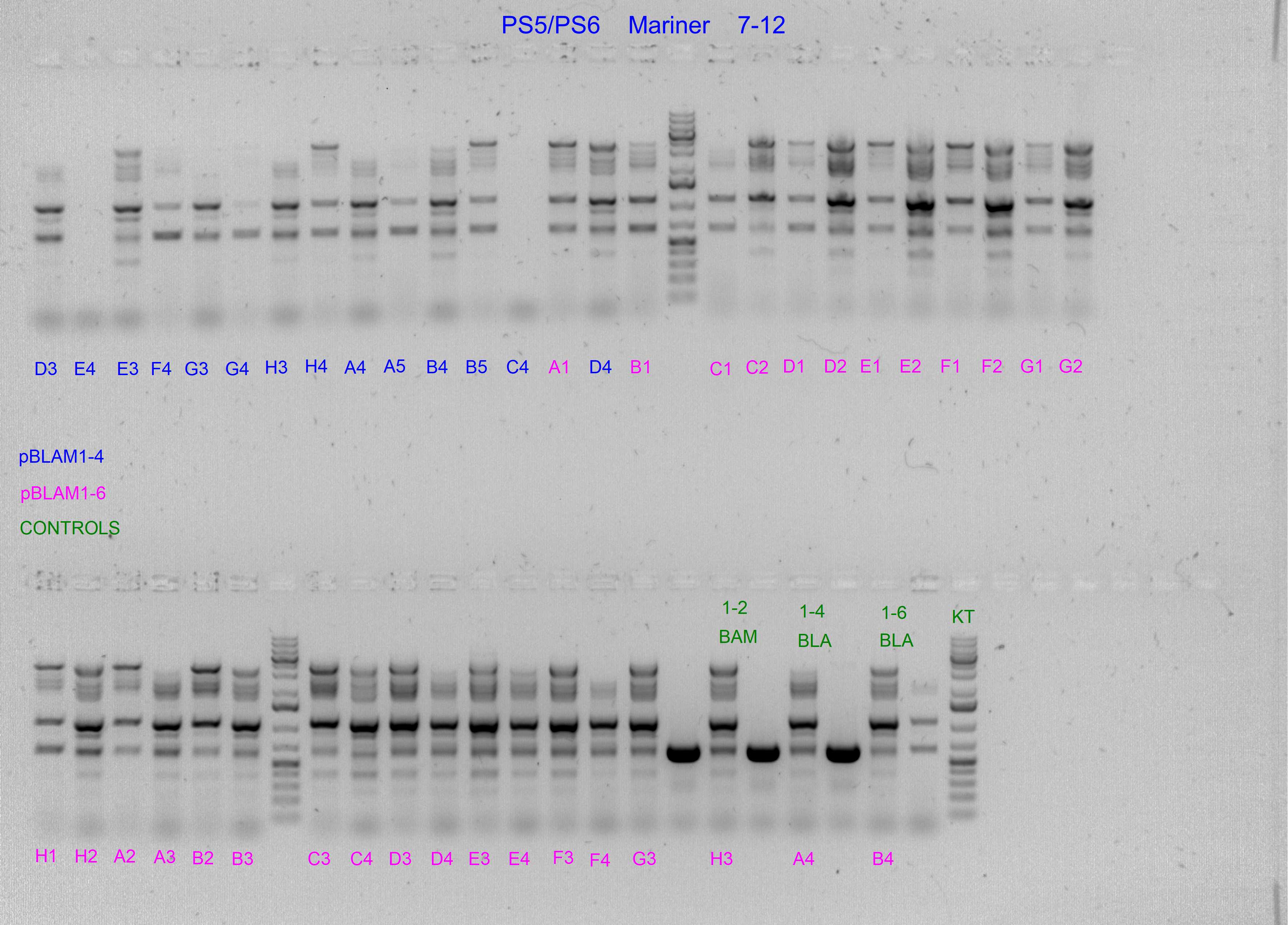
Spurious integration control with PS3/PS4 and PS5/PS6 SEVA oligonucleotides
The following template.csv was used for the OT-2 script in to prepare each mastermix with either PS3/PS4 or PS5/PS6 primer pairs and transfer cultures from Master 96-well plate
Gels after PCR protocol in thermocycler
Cargo integration control with PSMCS and either ME-O-Km-R/ME-O-Sm-R or ME-O-Gm-R
The following template.csv was used for the OT-2 script in to prepare the mastermix and transfer cultures from Master 96-well plate
We have discarted the first 3 columns because they came as negative (without integration) in the spurious PCRs.
Gels after PCR protocol in thermocycler
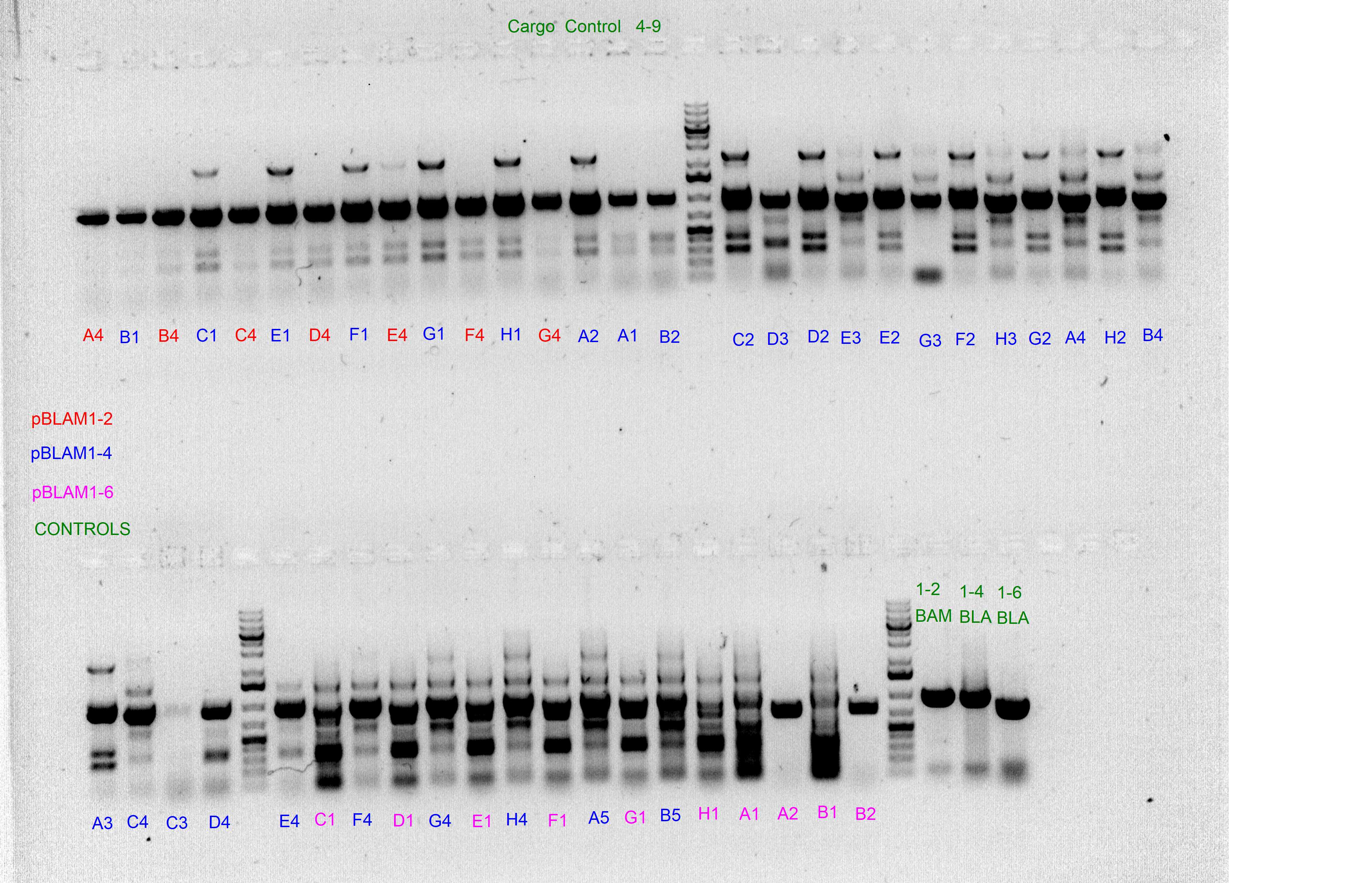
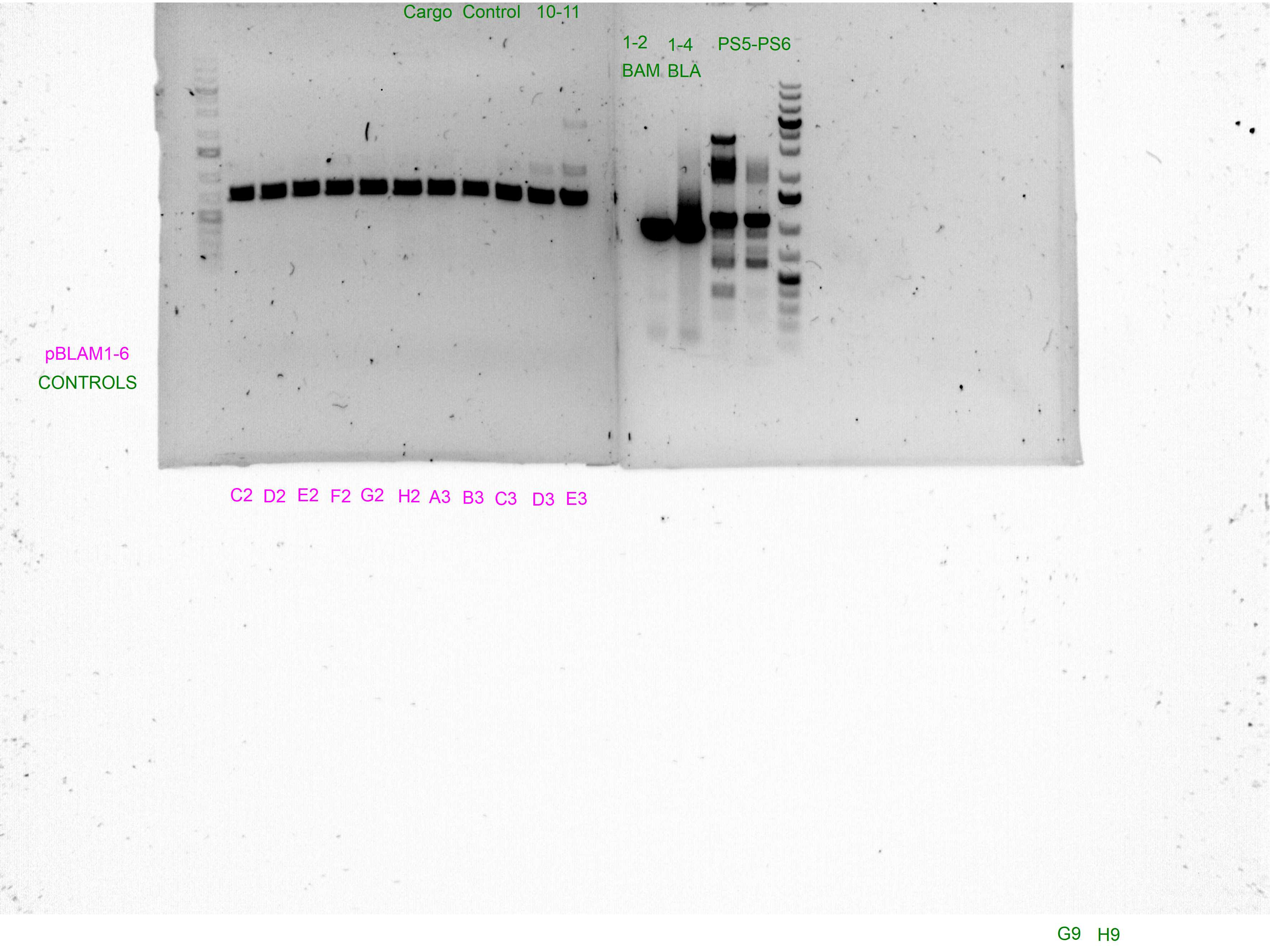
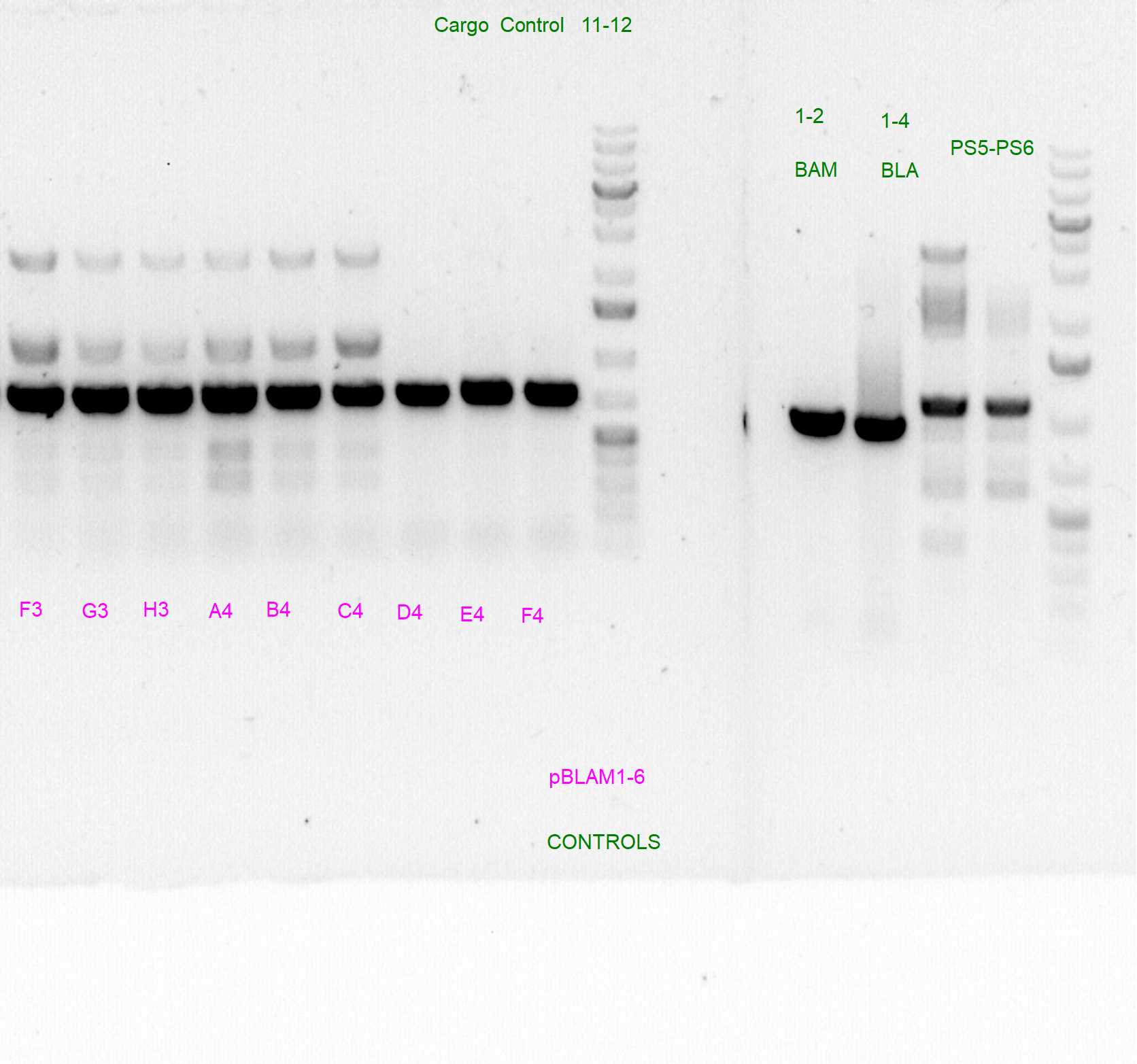
Arbitrary PCRs
Arbitrary PCR#1
The following three different template.csv were used (one for each primer pair) in three independent runs with the OT-2 script in using the same input and output plate to amplify the genomic context around each transposon cassette
Variables for source plate 1 arbitrary PCR 1: Variables-PCRs-OT.csv
Variables for source plate 2 arbitrary PCR 1: Variables-PCRs-OT.csv
Variables for source plate 3 arbitrary PCR 1: Variables-PCRs-OT.csv
Arbitrary PCR#2
The following three different template.csv were used (one for each primer pair) in three independent runs with the OT-2 script in using the Arbitrary PCR#1 plate as input and the same output plate to amplify the genomic context around each transposon cassette
Variables for source plate 1 arbitrary PCR 2: Variables-PCRs-OT.csv
Variables for source plate 2 arbitrary PCR 2: Variables-PCRs-OT.csv
Variables for source plate 3 arbitrary PCR 2: Variables-PCRs-OT.csv
Gel results for arbitrary PCRs
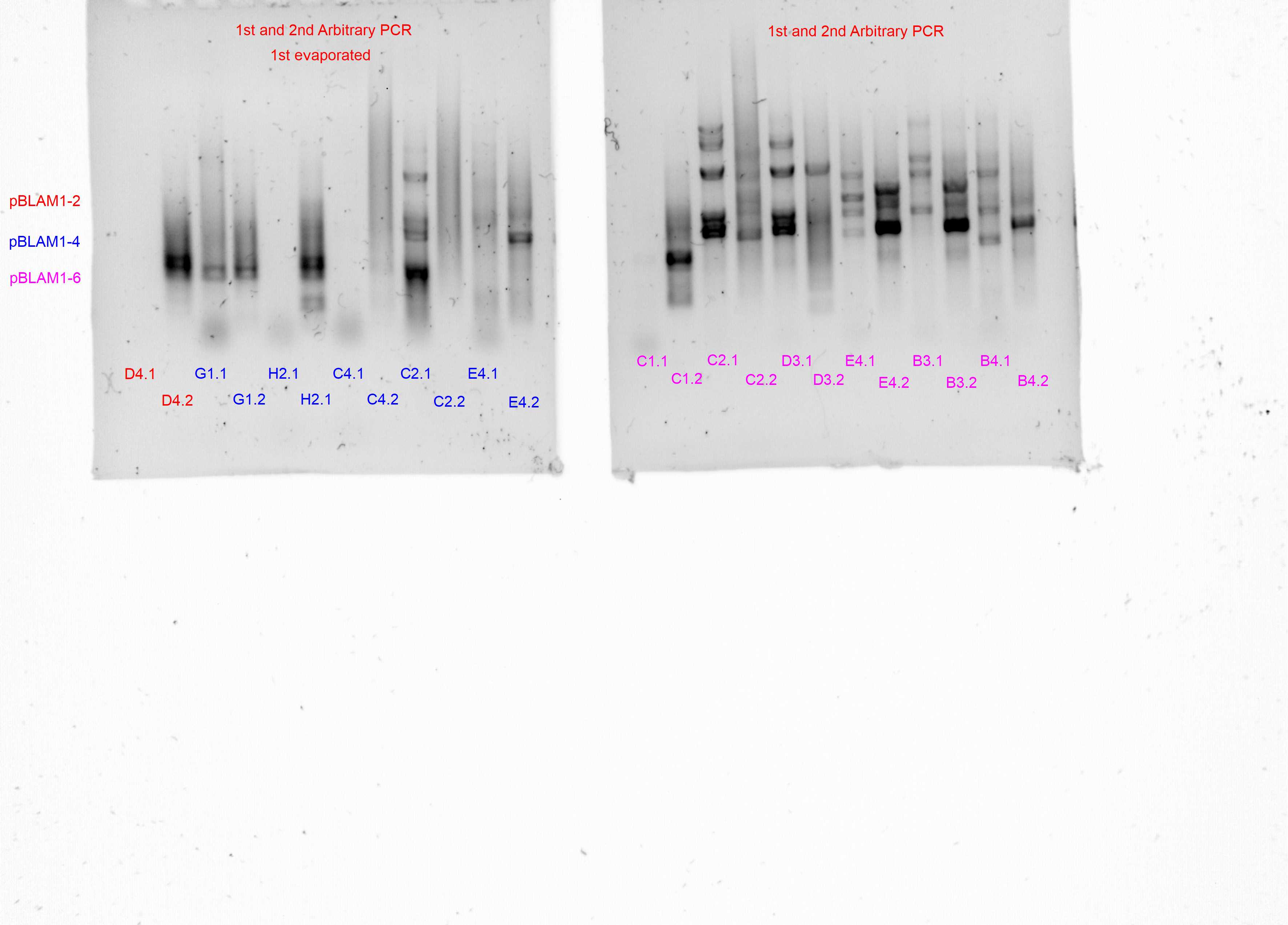
Sequencing and annotation of arbitrary PCR#2
The following files were used:
alignment_and_annotation_blastn.py Pseudomonas_putida_KT2440_110.fna Pseudomonas_putida_KT2440_110.csv sequencing_results.zip

