Getting started with Micro-Meta App Tutorial
David Grunwald, Alessandro Rigano, Mathias Hammer, Joel Ryan, Claire M. Brown, Caterina Strambio De Castillia
Abstract
For quality, interpretation, reproducibility, and sharing value, microscopy images should be accompanied by detailed descriptions of the conditions that were used to produce them. Micro-Meta App is an intuitive, highly interoperable, open-source software tool that was developed in the context of the 4D Nucleome (4DN) consortium and is designed to facilitate the extraction and collection of relevant microscopy metadata as specified by the recent 4DN-BINA-OME tiered-system of Microscopy Metadata specifications. In addition to substantially lowering the burden of quality assurance, the visual nature of the Micro-Meta App makes it particularly suited for training purposes.
In this protocol and in the associated videos (https://youtube.com/playlist?list=PLd2SsBj4SIsh7eE1M7l4v5_EsjUQcsfb0) you will learn how to get started using the Micro-Meta App for documenting your imaging experiments.
Want to learn more?
Please watch Video 1 of the Tutorial Series and read the publications listed in the References section of this protocol to learn more about the Micro-Meta App and the underlying 4DN-BINA-OME tiered system of Microscopy Metadata specifications.
Before start
In order to participate in this tutorial, you will need some prerequisites and materials that are described in the Materials section of this protocol. Materials section of this protocol.
In particular, before you start you will need to download the following example metadata files:
- Microscope.JSON file: A file that contains a description of the hardware components of an example microscope.
- Settings.JSON file: A file that contains a description of the settings that were used to acquire the example image data file.
- Raw-image data file: An image data file that was acquired using the hardware specifications and acquisition settings described in the Microscope.JSON and Settings.JSON files.
Steps
Before the tutorial
Video introduction
#尊敬的用户,由于网络监管政策的限制,部分内容暂时无法在本网站直接浏览。我们已经为您准备了相关原始数据和链接,感谢您的理解与支持。
<iframe width="560" height="315" src="https://www.youtube.com/embed/lNumFns3yMQ" title="YouTube video player" frameborder="0" allow="accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
#尊敬的用户,由于网络监管政策的限制,部分内容暂时无法在本网站直接浏览。我们已经为您准备了相关原始数据和链接,感谢您的理解与支持。
<div style="padding:56.25% 0 0 0;position:relative;"><iframe src="https://player.vimeo.com/video/604291798?h=eee347a8ee&badge=0&autopause=0&player_id=0&app_id=58479" frameborder="0" allow="autoplay; fullscreen; picture-in-picture" allowfullscreen style="position:absolute;top:0;left:0;width:100%;height:100%;" title="Introduction to the use of Micro-Meta App."></iframe></div><script src="https://player.vimeo.com/api/player.js"></script>
Tutorial - 1 - Download and Install Micro-Meta App
Download Micro-Meta App
- Follow the instructions in this step and in Video 3 of the tutorial series tutorial series to download and install the stand-alone version of the Micro-Meta App.
Click on this link to access the Download page
Download and save ZIP file
- Once you are on the download page please navigate to the bottom page and identify the ZIP file that is best suited for your operating system.
- "Right-click" on the link and select "Save Link As..."
- Save the ZIP file to the "MMA Tutorial" folder you have created to save the example metadata files (see the instructions that are found in the Materials section of this protocol).
Unarchive the ZIP file and launch Micro-Meta App
- Unarchive the ZIP file by following the instructions that are appropriate for your operating system.
Note
IMPORTANT NOTE : If you are planning to use Micro-Meta App on Mac OS, DO NOT USE the native Mac OS unarchiver utility to unzip the ZIP file. Instead, we strongly recommend that you use If you are planning to use Micro-Meta App on Mac OS, DO NOT USE the native Mac OS unarchiver utility to unzip the ZIP file. Instead, we strongly recommend that you use The Unarchiver.app..For additional instructions please refer to the Materials section of this protocol.In order to use In order to use The Unarchiver.app instead of the native unarchiver Mac OS utility, follow these instructions instead of the native unarchiver Mac OS utility, follow these instructionsLocate the downloaded micro-meta-app-electron-XXX-mac.zip file"Right-click" (do not double click) on the ZIP fileClick on "Open-with" Select "The Unarchiver.app" from the menu
Launch Micro-Meta App
- Launch the Micro-Meta App by following the instructions that are appropriate for your operating system.
Note
IMPORTANT NOTE : If you are planning to use Micro-Meta App on Mac OS, please use the special instructions below when you launch the app for the first time.Locate the icon corresponding to the unzipped Micro-Meta App executable file."Right-click" (do not double click) on the executable file and select "Open".The system will give you an error message, click on "Cancel"Right-click" (do not double-click) on the executable file and select "Open", again.The system will give you an error and request if you want to continue, click on "OK".From this point forward, it will be possible to launch the app by double-clicking on the executable file icon.
Tutorial - 2 - Manage Instrument
Manage Instrument
In this section of the tutorial, you will learn how to use the Manage Instrument functionality of Micro-Meta App to open an existing Microscope.JSON file and to create a simple new Microscope.JSON file from scratch.
To learn more about the Tier system of Microscopy Metadata used as the basis of Micro-Meta App please read the following publication:
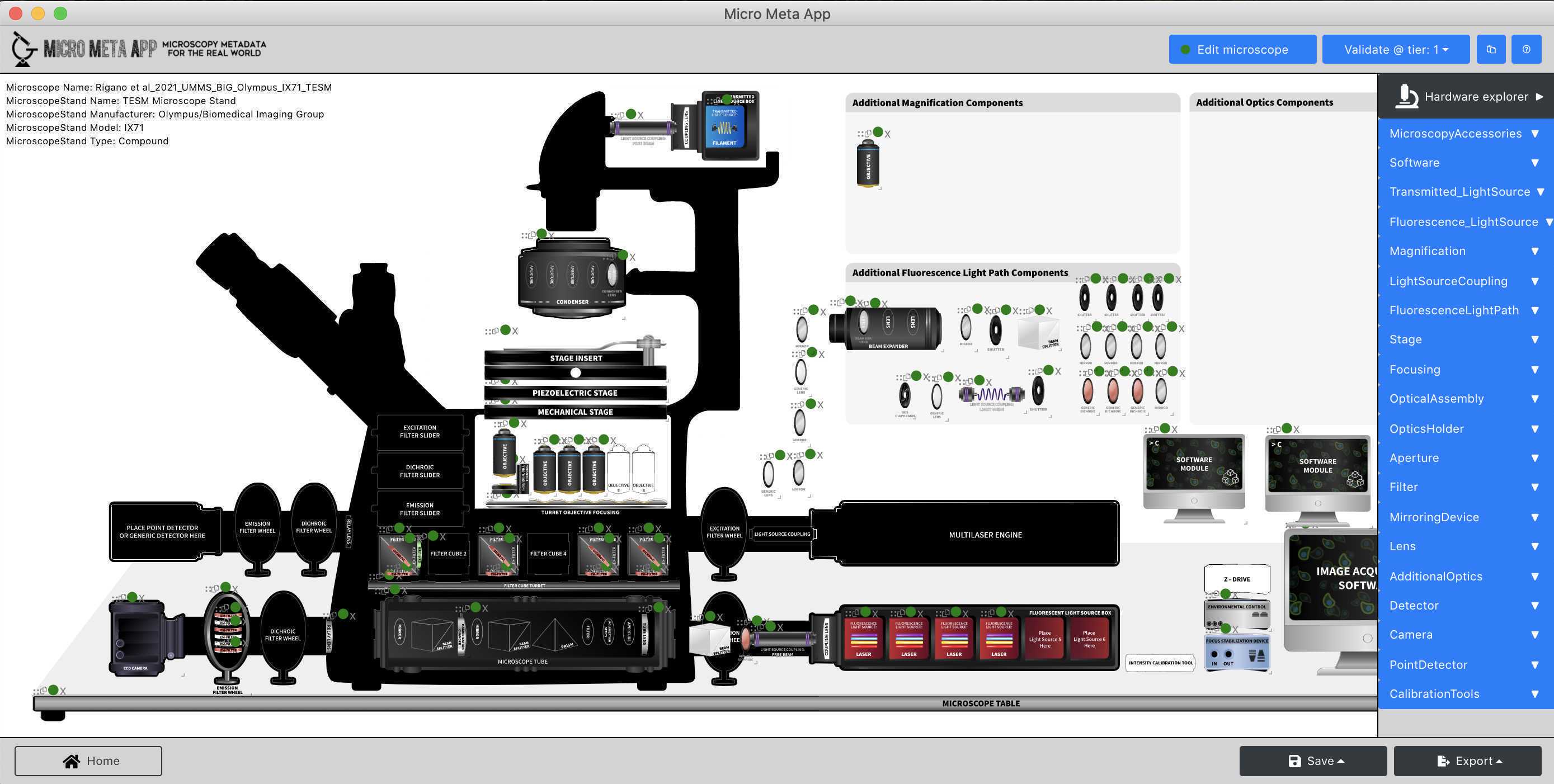
Open and inspect an existing Microscope.JSON file
In this part of the tutorial, you will learn how to open and inspect an existing Microscope.JSON file to evaluate its content and decide whether it needs further editing.
To get started, follow these steps:
- Launch Micro-Meta App as described
- Click on "Continue".
- Click on “Manage Instrument” .
- Click on "Tier 3 , " .
- Click on “Import from file” and select the example Microscope.JSON file downloaded from Zenodo as described in the MATERIALS section and above , and click on "Continue".
- Follow the instructions in Video 4 of the Tutorial series Video 4 of the Tutorial series to explore the hardware specifications metadata contained in the example Microscope.JSON file.
Create a new Microscope.JSON file from scratch
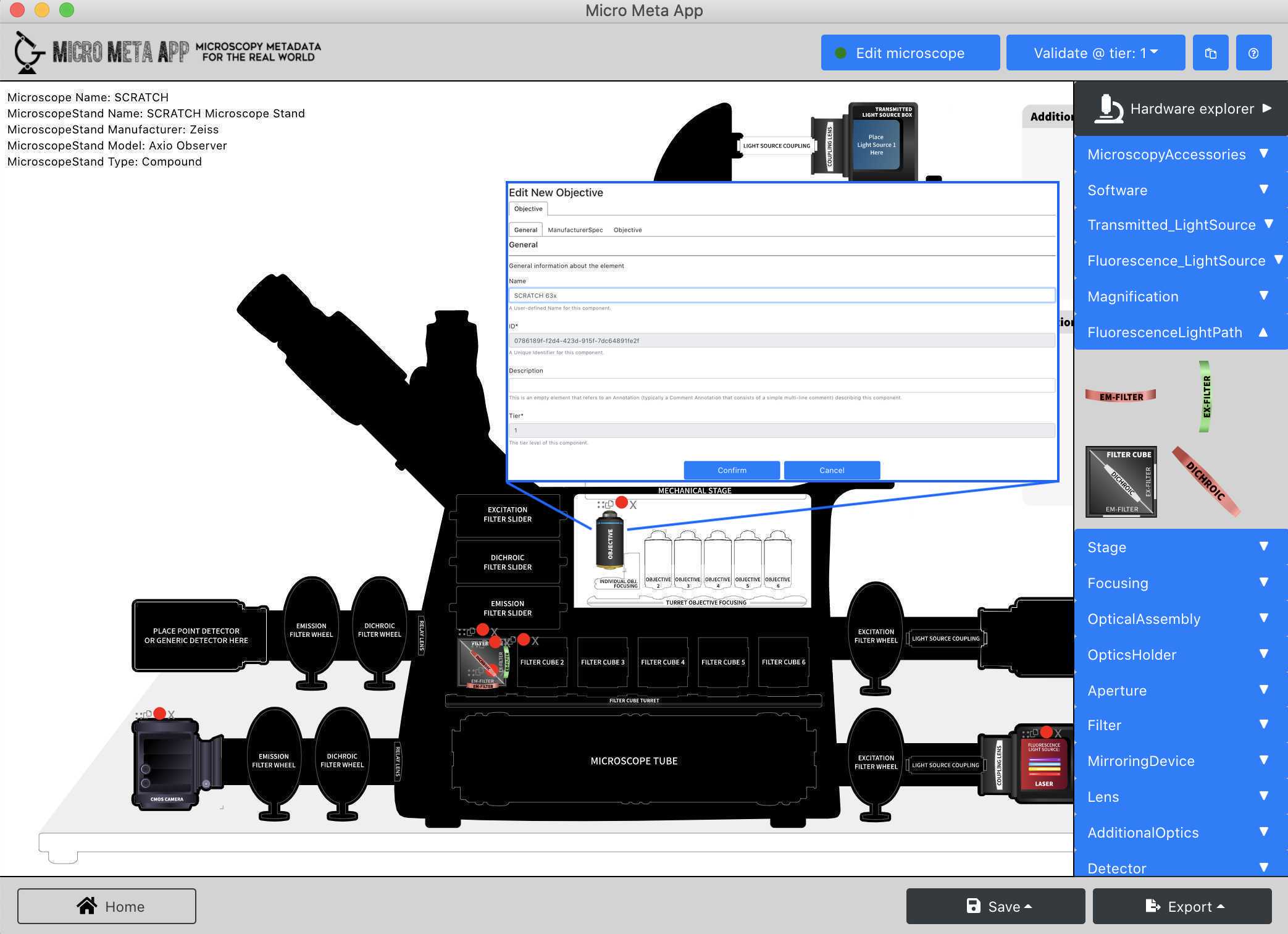
In this part of the tutorial, you will learn how to create a new Microscope.JSON file that describes a Tier 1 microscope that is only used for qualitative or simple quantitative experiments.
To get started follow the steps hereinafter:
-
Launch Micro-Meta App as described
-
Click on "Continue".
-
Click on “Manage Instrument” .
-
Click on "Tier 1" .
-
Click on “Create Inverted from scratch” , and click on "Continue".
-
Follow the instructions in Video 5 of the tutorial series Video 5 of the tutorial series to add the following hardware components to the Microscope canvas and enter the hardware specifications listed in the specification files found following the links listed below: → Microscope Stand → Zeiss Axio-Observer
→ **Fluorescence Light Source/LED** → [Zeiss Colibri 5/7 LED](https://asset-downloads.zeiss.com/catalogs/download/mic/991e3533-433d-4964-8781-f3774afb558b/EN_product-info_Colibri-7_rel1-0.pdf) → **Magnification/Objective →** [Zeiss PlanApo 63x/1.4](https://www.micro-shop.zeiss.com/en/us/shop/objectives/420782-9900-799/Objective-Plan-Apochromat-63x-1.4-Oil-DIC-M27) → **Fluorescence Light Path/Filter Cube** - **Filter Set →** [Multiband filter set, optimized for GFP & DsRed](https://www.idex-hs.com/store/product-detail/gfp_dsred_a_000/s-000780?cat_id=products&node=optical_filter_sets) ○[Excitation Filter](https://www.idex-hs.com/store/product-detail/ff01_468_553_25/fl-004593)r [Emission Filter](https://www.idex-hs.com/store/product-detail/ff01_512_630_25/fl-004381)er [Dichroic beamsplitter](https://www.idex-hs.com/store/product-detail/ff493_574_di01_25x36/fl-006972)ter → **Detector →** [Andor iXon Life 888 camera](https://andor.oxinst.com/assets/uploads/products/andor/documents/andor-ixon-life-emccd-specifications.pdf)
Tutorial - 3 - Manage Settings
Manage Settings
In this section of the tutorial, you will learn how to use the Manage Settings functionality of theMicro-Meta App to open an existing Settings.JSON file, inspect its content and add a missing transmitted light Channel.
Open and inspect an existing Settings.JSON file
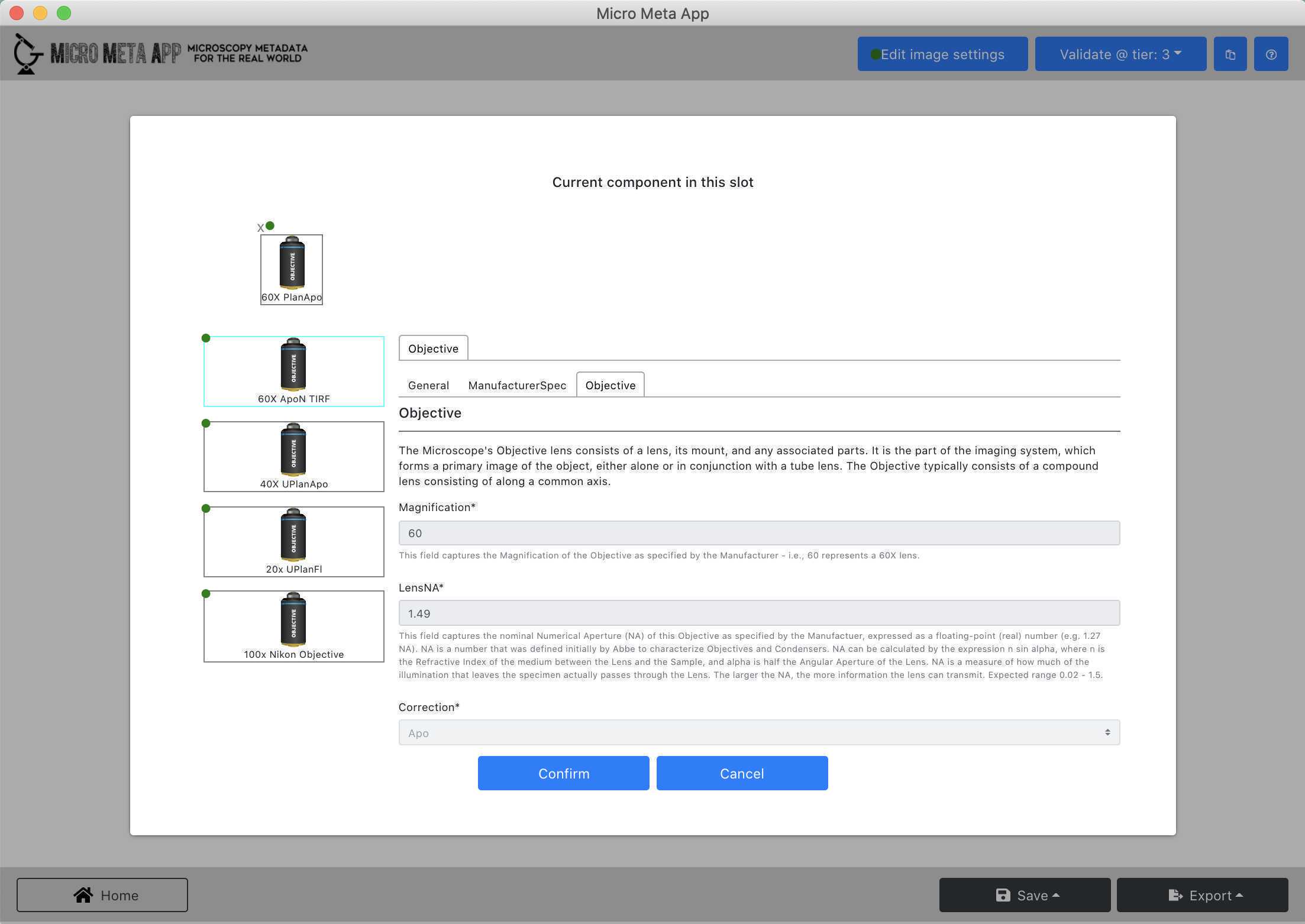
In this part of the tutorial, you will learn how to open and inspect an existing Settings.JSON file to evaluate its content and decide whether it needs further editing.
- Launch Micro-Meta App as described
- Click on "Continue".
- Click on “Manage Settings .
- Click on "Tier 3" .
- Click on “Import from file”, select the example Microscope.JSON file downloaded from Zenodo as described in the MATERIALS section and above , and click "Continue".
- Click on “Import from file” , select the example image-data file downloaded from Zenodo as described in the MATERIALS section and above , and click "Continue".
- Click on “Import from file” , select the example Settings.JSON file downloaded from Zenodo as described in the MATERIALS section and above , and click "Continue".
- Follow the instructions in Video 6 of the tutorial series Video 6 of the tutorial series to explore the image acquisition metadata contained in the example Settings.JSON file.
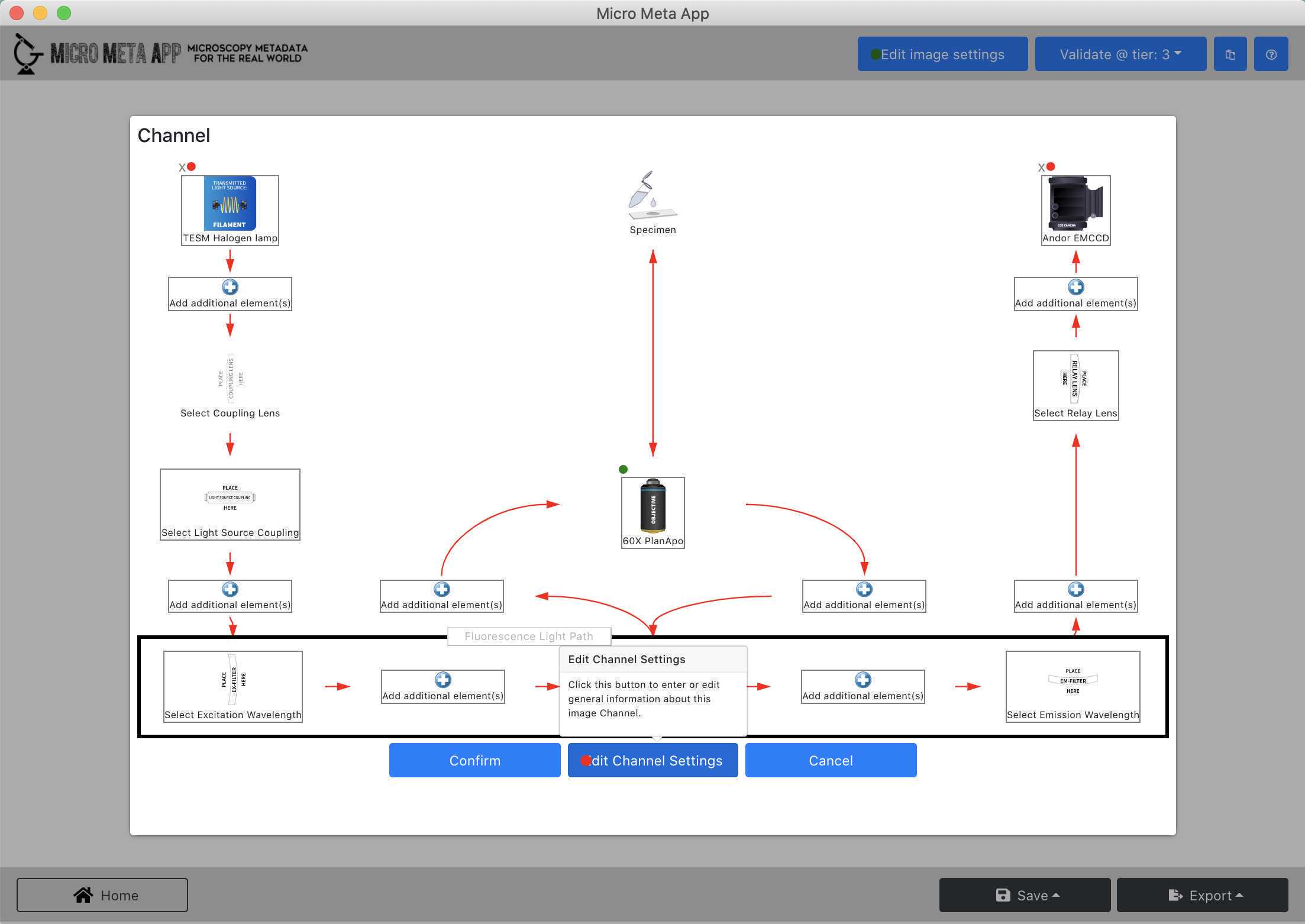
Add an additional transmitted light Channel to an existing Settings.JSON file
- Follow the steps described here
- Click on "Edit Channels".
- Click on the "+" button, click on "Channel 3" , and click on "Edit selected".
- Follow the instructions in Video 7 of the tutorial series Video 7 of the tutorial series to enter information about a new transmittance light Channel in an existing Settings.JSON file.
After the tutorial
Provide feedback
After testing the Micro-Meta App, we would love to have your feedback.
- Please fill in the survey by clicking here
Want to learn more?
Please read the publications and consult the resources listed below to learn more about Micro-Meta App, the underlying 4DN-BINA-OME tiered system of Microscopy Metadata specifications, and related metadata collection tools.
Community-driven 4DN-BINA-OME (NBO) Microscopy Metadata Specifications
To learn more about the 4DN-BINA-OME tiered system of Microscopy Metadata specifications that underlies the Micro-Meta App please consult the resources below:
Software
| Value | Label |
|---|---|
| 4DN-BINA-OME Microscopy Metadata Specifications | NAME |
| https://github.com/WU-BIMAC/NBOMicroscopyMetadataSpecs | REPOSITORY |
| Mathias Hammer, Alessandro Rigano, Caterina Strambio-De-Castillia | DEVELOPER |
| https://zenodo.org/record/4710731 | LINK |
| v2.01 | VERSION |
Micro-Meta App
To learn more about the Micro-Meta App please consult the resources below:
Software
| Value | Label |
|---|---|
| Micro-Meta App | NAME |
| MacOS, Windows | OS_NAME |
| https://github.com/WU-BIMAC/MicroMetaApp-Electron | REPOSITORY |
| Alessandro Rigano, Caterina Strambio-De-Castillia | DEVELOPER |
| https://github.com/WU-BIMAC/MicroMetaApp-Electron/releases/latest | LINK |
| v1.6.19-beta | VERSION |
Other metadata collection tools
To learn more about other interoperable metadata collection tools please consult the resources below:
Software
| Value | Label |
|---|---|
| MethodsJ2 | NAME |
| https://github.com/ABIF-McGill/MethodsJ2 | REPOSITORY |
| Joel Rigano, Claire M. Brown | DEVELOPER |
Get more involved
If you want to get more involved and help us with developing or testing Micro-Meta App please join us by joining the QUAREP-LiMi Working Group WG7 on Metadata by clicking on the link below:

