Generation of ‘semi-guided’ cortical organoids with complex neural oscillations
Michael Q. Fitzgerald, Tiffany Chu, Francesca Puppo, Rebeca Blanch, Miguel Chillón, Shankar Subramaniam, Alysson R. Muotri

















Extended
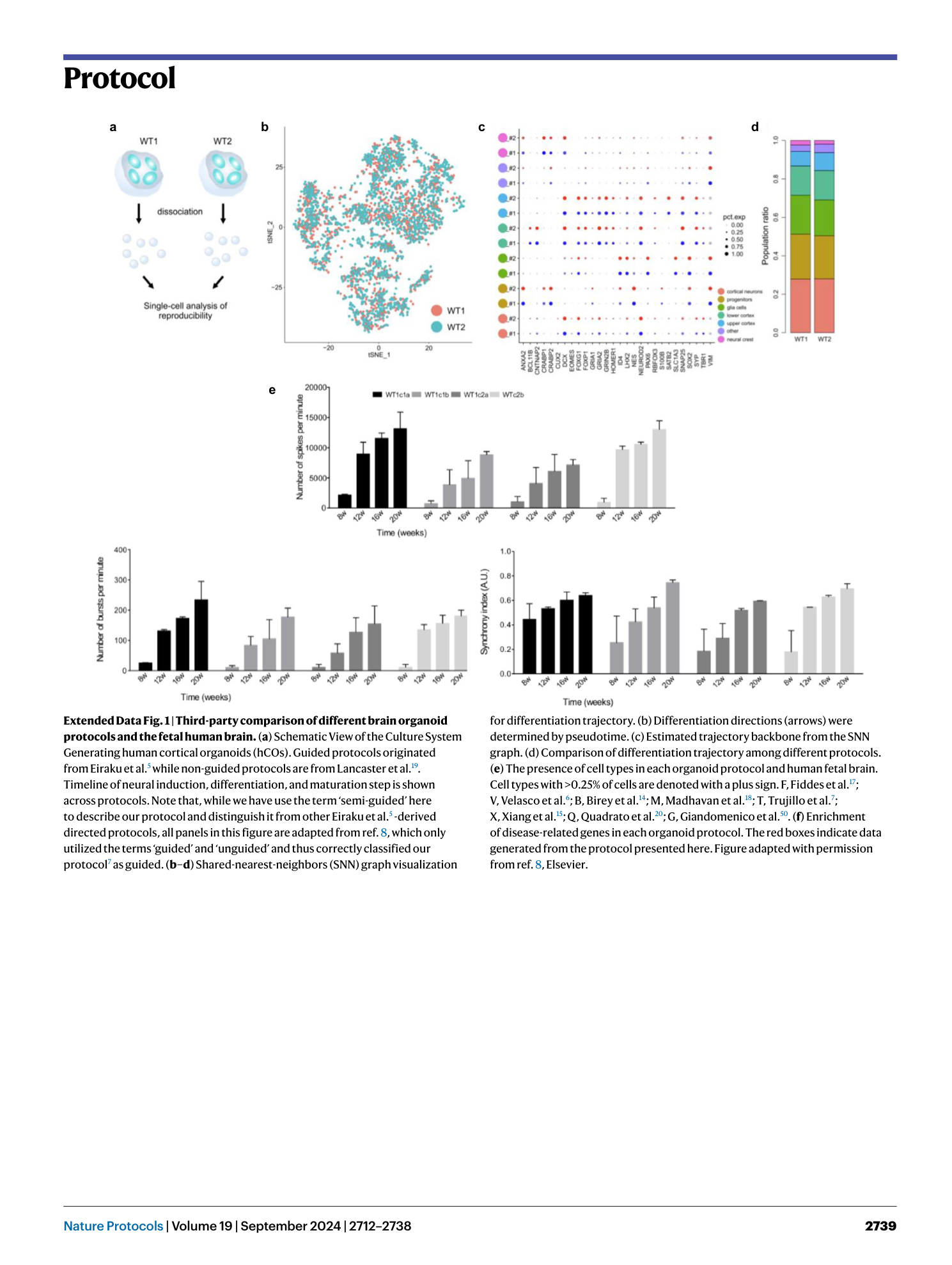
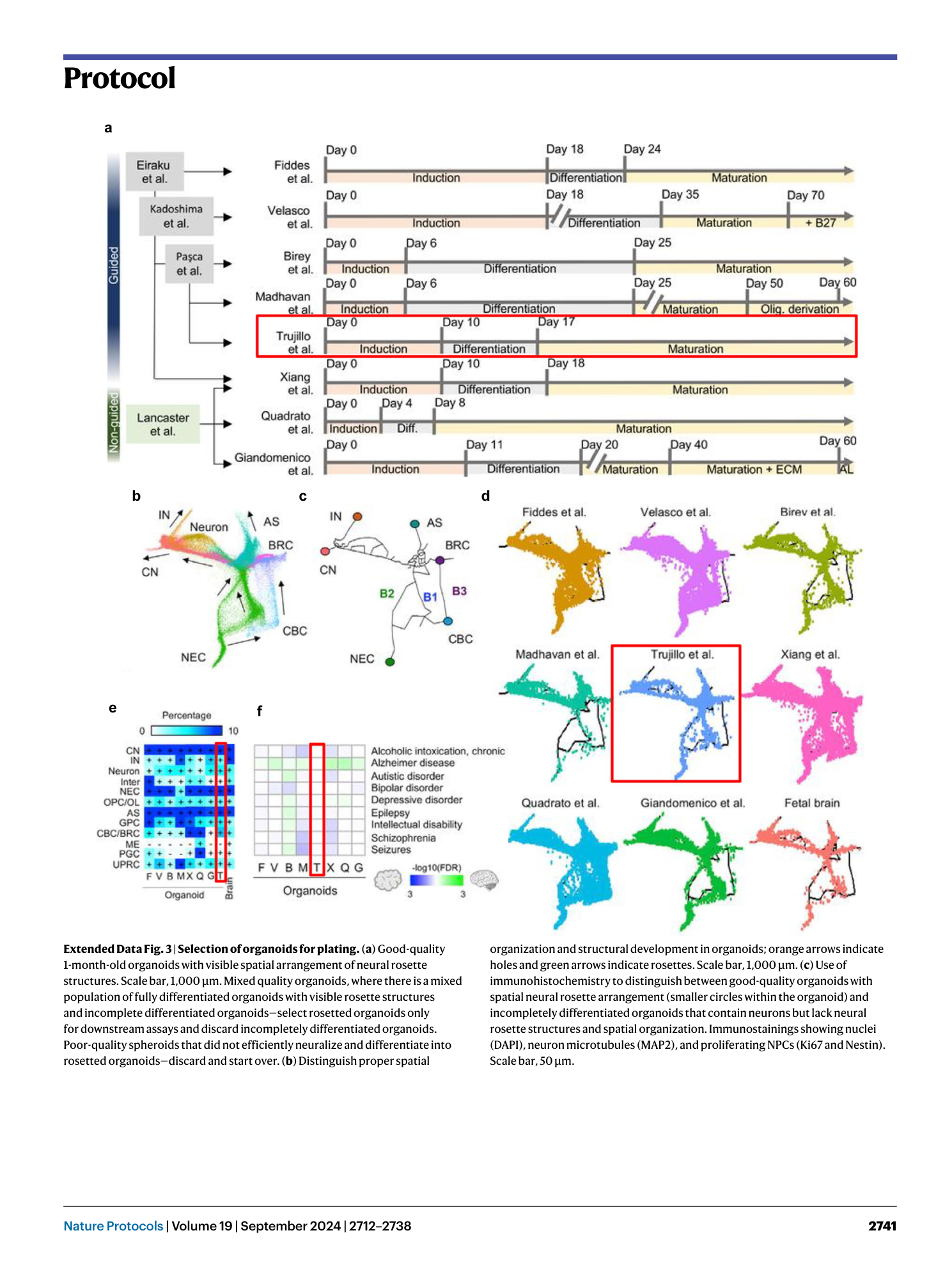
Extended Data Fig. 1 Third-party comparison of different brain organoid protocols and the fetal human brain.
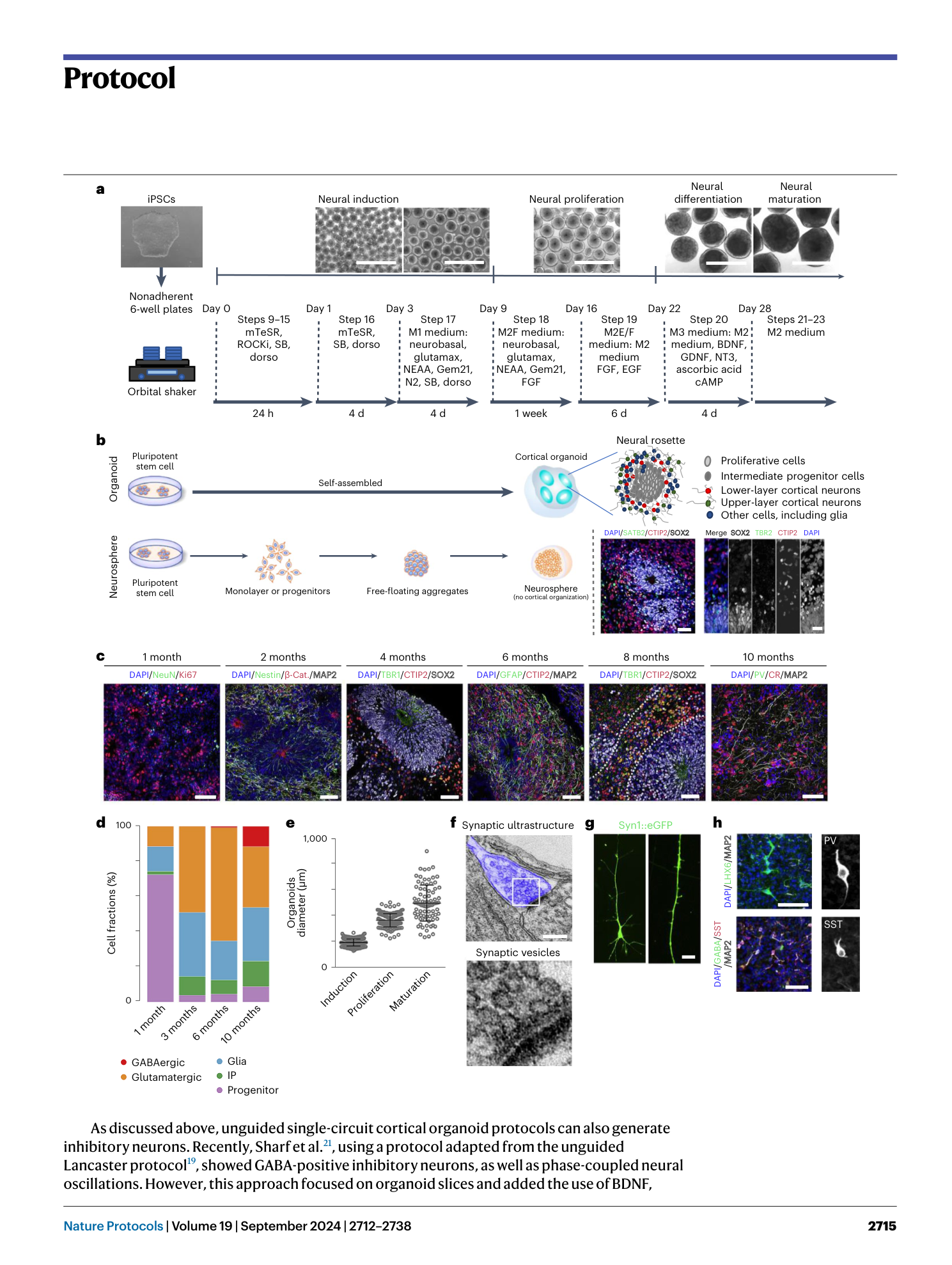
( a ) Schematic View of the Culture System Generating human cortical organoids (hCOs). Guided protocols originated from Eiraku et al. 5 while non-guided protocols are from Lancaster et al. 19 . Timeline of neural induction, differentiation, and maturation step is shown across protocols. Note that, while we have use the term ‘semi-guided’ here to describe our protocol and distinguish it from other Eiraku et al. 5 -derived directed protocols, all panels in this figure are adapted from ref. 8 , which only utilized the terms ‘guided’ and ‘unguided’ and thus correctly classified our protocol 7 as guided. ( b–d ) Shared-nearest-neighbors (SNN) graph visualization for differentiation trajectory. (b) Differentiation directions (arrows) were determined by pseudotime. (c) Estimated trajectory backbone from the SNN graph. (d) Comparison of differentiation trajectory among different protocols. ( e ) The presence of cell types in each organoid protocol and human fetal brain. Cell types with >0.25% of cells are denoted with a plus sign. F, Fiddes et al. 17 ; V, Velasco et al. 6 ; B, Birey et al. 14 ; M, Madhavan et al. 18 ; T, Trujillo et al. 7 ; X, Xiang et al. 15 ; Q, Quadrato et al. 20 ; G, Giandomenico et al. 50 . ( f ) Enrichment of disease-related genes in each organoid protocol. The red boxes indicate data generated from the protocol presented here. Figure adapted with permission from ref. 8 , Elsevier.
Extended Data Fig. 2 Molecular and functional reproducibility of cortical organoids.
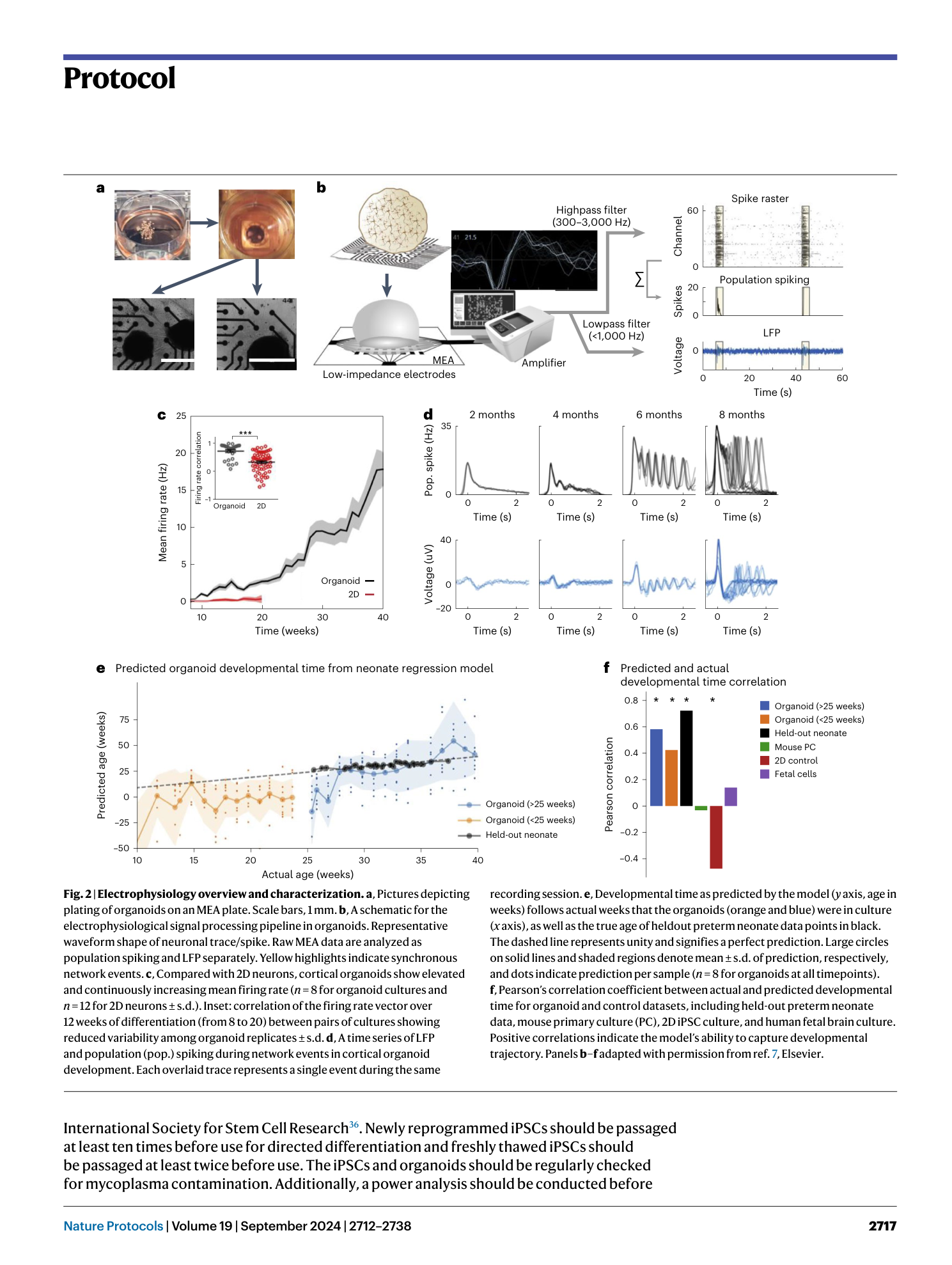
( a ) Schematic showing the single-cell approach performed to assess the reproducibility of organoid generation using different iPSC lines (WT1 and WT2). ( b ) tSNE plot of single-cell mRNA sequencing data from two 6-month-old organoids. ( c ) Expression of gene markers for various cell types both batches. ( d ) Population ratio of each cluster by replicate. ( e ) Consistent and reproducible development of electrical activity in organoids over time across four cell lines, bars represent mean ± s.e.m (n = 8, independent experiments performed in duplicates using two clones of an iPSC line).
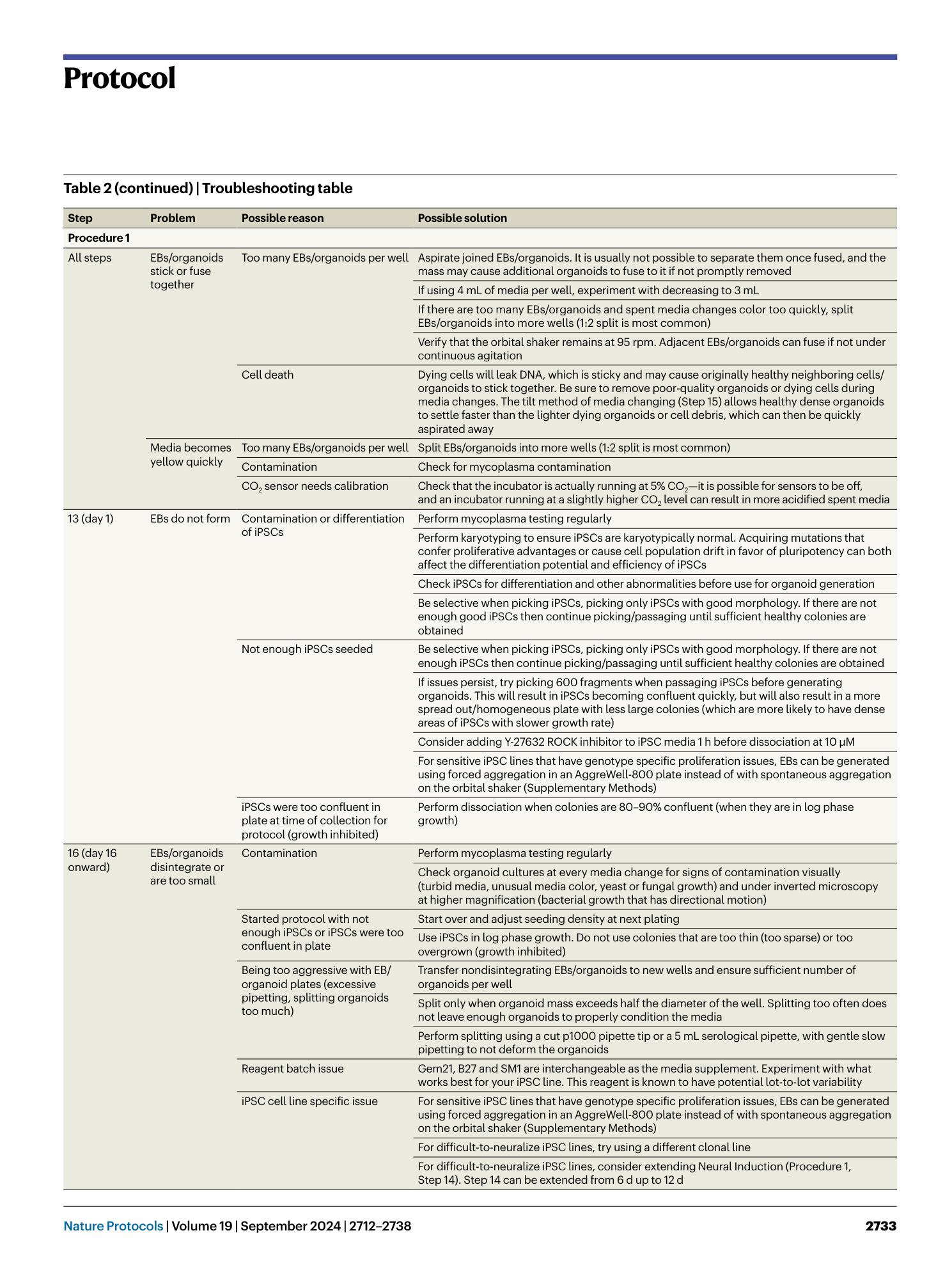
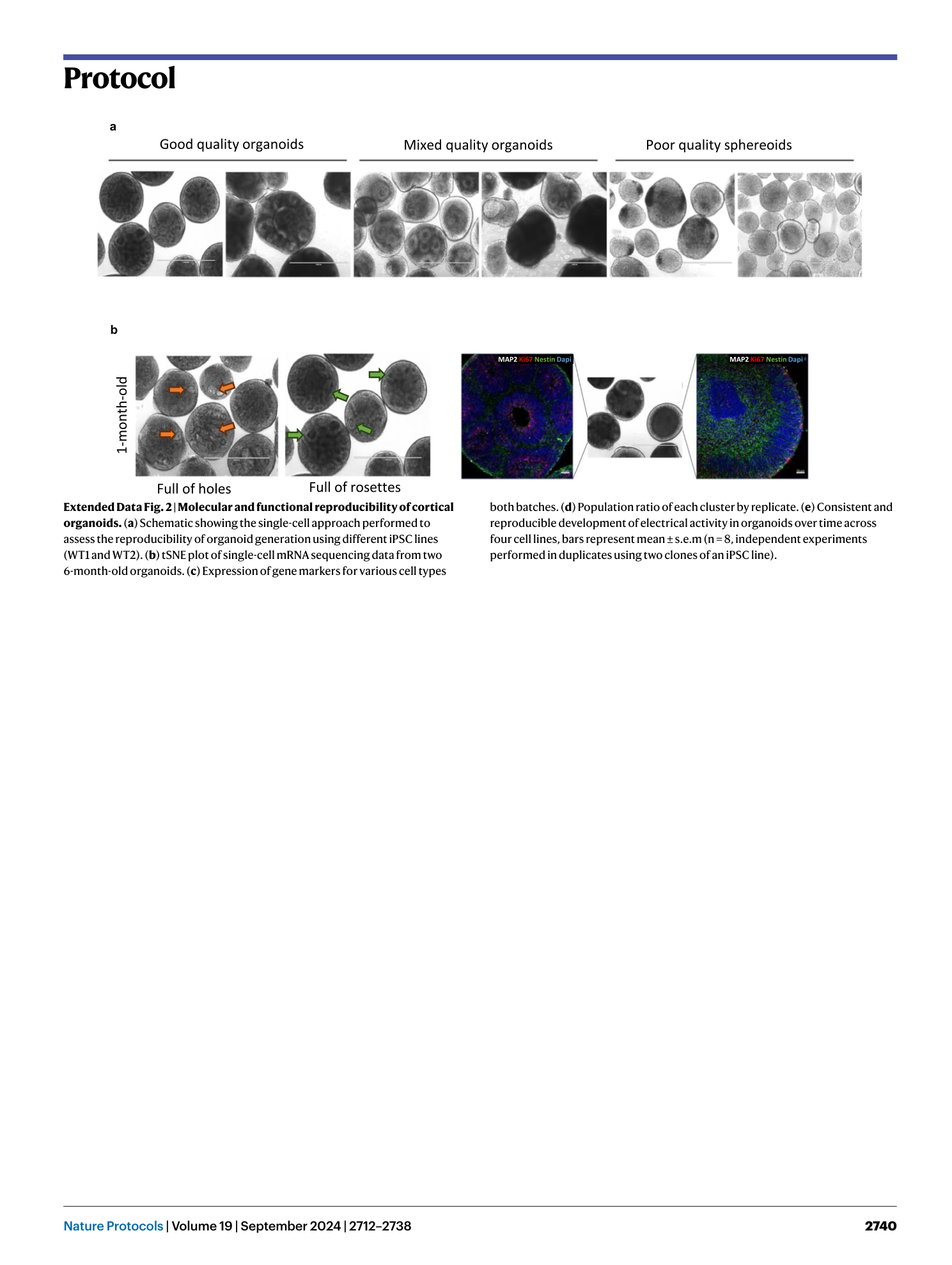
Extended Data Fig. 3 Selection of organoids for plating.
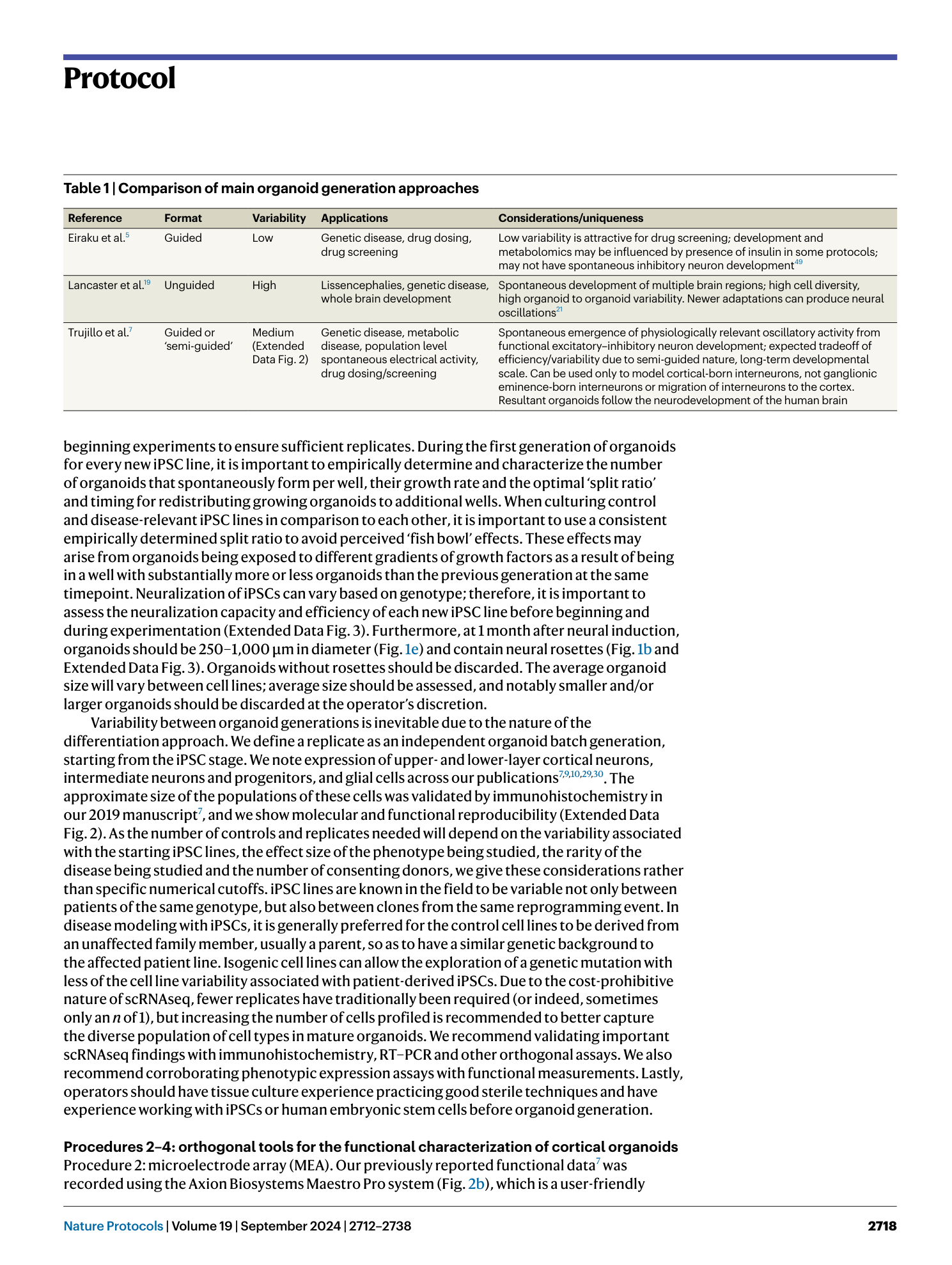
( a ) Good-quality 1-month-old organoids with visible spatial arrangement of neural rosette structures. Scale bar, 1,000 μm. Mixed quality organoids, where there is a mixed population of fully differentiated organoids with visible rosette structures and incomplete differentiated organoids—select rosetted organoids only for downstream assays and discard incompletely differentiated organoids. Poor-quality spheroids that did not efficiently neuralize and differentiate into rosetted organoids—discard and start over. ( b ) Distinguish proper spatial organization and structural development in organoids; orange arrows indicate holes and green arrows indicate rosettes. Scale bar, 1,000 μm. ( c ) Use of immunohistochemistry to distinguish between good-quality organoids with spatial neural rosette arrangement (smaller circles within the organoid) and incompletely differentiated organoids that contain neurons but lack neural rosette structures and spatial organization. Immunostainings showing nuclei (DAPI), neuron microtubules (MAP2), and proliferating NPCs (Ki67 and Nestin). Scale bar, 50 μm.
Supplementary information
Supplementary Information
Supplementary Methods.
Supplementary Video 1
Representative movies of calcium activity in GCaMP-expressing cortical organoids
Supplementary Video 2
Representative movies of calcium activity in OGB1-labeled 2D cultures of iPSC-derived neurons. 2D cortical neurons are 4 and 5 months old, respectively.
Supplementary Video 3
Representative movies of calcium activity from neurons from OGB1- labeled organoids plated on imaging plates.

