Generating Mutant Renal Cell Lines Using CRISPR Technologies
Nuria Perretta-Tejedor, Grace Freke, Marian Seda, David. A. Long, Dagan Jenkins
Abstract
Gene editing using the CRISPR/Cas9 system is an extremely efficient approach for generating mutations within the genomic DNA of immortalized cell lines. This procedure begins with a straightforward cloning step to generate a single plasmid encoding the Cas9 enzyme as well as a synthetic guide RNA (sgRNA) which is selected to target specific sites within the genome. This plasmid is transfected into cells either alone, in order to generate random insertion-deletion alleles (“indels”) at the desired locus via the nonhomologous end-joining pathway, or in conjunction with a homology-directed repair template oligonucleotide to generate a specific point mutation. Here we describe a procedure to perform gene editing in IMCD3 and HEK293 cells and to subsequently isolate clonal cell lines carrying mutations of interest.
Before start
The methods presented below should be used to generate random indels by NHEJ. A variation of this protocol can be used to generate a defined single base change mutation (or if required, substitution, insertion, or deletion of a few nucleotides) by HDR ( see Note 1 ). For simplicity, the following methods describe the design and use of one sgRNA ( see Note 2 ; Fig. 2).

Steps
3.1 sgRNA Design
Find your gene of interest within the relevant species on the Ensembl genome browser (www.ensembl.org). For HEK293 search the human genome, and for IMCD3 search the mouse genome.
Select a 23–500-nucleotide (nt) genomic region that you wish to edit ( see Note 3 ).
Open the CRISPOR design tool (crispor.tefor.net). Select the appropriate target genome and paste your sequence into the search box. Submit the search. The online tool will produce a ranked list of all possible sgRNAs within the genomic sequence ( see Note 4 ).
Identify a suitable sgRNA sequence from the displayed list ( see Note 5 ).
Take the 20 nt sgRNA sequence (without the PAM). If its first nucleotide is not a “G,” then add a “G” at the 5′ end. This sequence will form the foundations of oligo 1.
Find the reverse complement of the sgRNA sequence (including the extra “G” nucleotide, if added). This will form the foundations of oligo 2.
To the 5′ end of oligo 1, insert nucleotides “CACC.” Design of oligo 1 is now complete.
To the 5′ end of oligo 2, insert nucleotides “AAAC.” Design of oligo 2 is now complete.
Order oligos 1 and 2 as 5′ phosphorylated oligos.
Design a pair of PCR primers flanking the sgRNA genomic target site ( see Note 6 ).
3.2 Cloning of sgRNA into Plasmid pX330
3.2.1 Annealing of Oligos 1 and 2
Resuspend each oligo (ordered in Section 3.1, step 9 ) to a concentration of 100micromolar (µM) in Milli-Q water.
In a 0.2 mL PCR tube, combine 1µL , 1µL, and 8µL.
Anneal in a thermocycler using the following cycling conditions: 95°C for 0h 5m 0s, ramp down to 25°C at 5°C/min.
3.2.2 pX330 Linearization
On ice, prepare a 50µL reaction containing 100ng , 5µL , 4µL , and Milli-Q water to a final volume of 50µL.
Incubate for 2h 0m 0s at 37°C.
3.2.3 Ligation of Annealed Oligos into pX330
Dilute annealed oligos (from Section 3.2.1, step 11 ) 1:200 in Milli-Q water.
Prepare a 10µL (from Section 3.2.2, step 12 ), 1µL, 1µL, 1µL, and Milli-Q water to a final volume of 10µL.
Incubate for at least 1h 0m 0s at Room temperature, or at 4°C.
3.2.4 Transformation of Competent Cells with the Ligation Mix and Isolation of Plasmid DNA
Thaw E. coli JM109 competent cells On ice until just thawed.
Add 5µL (from Section 3.2.3, step 13 ) to the tube of competent cells. Mix by flicking the tube 4–5 times (do not mix by pipetting) and incubate cells for 0h 30m 0s On ice.
Heat shock the cells for 0h 0m 45s in a water bath at exactly 42°C; do not shake. Immediately return the tube to ice for 0h 2m 0s.
Add 450µL to the tube and incubate for 1h 0m 0s at 37°C with shaking.
Plate 100µL onto an LB agar Amp plate.
Incubate plate at 37°C.
The next day, inspect the plate for colony growth. Pick two or three colonies to check for correct insertion of the sgRNA. Inoculate a single colony into a 5mL . Incubate cultures at 37°C with shaking.
Isolate plasmid DNA from the cultures using a QIAprep Spin Miniprep kit, according to the manufacturer’s instructions.
Verify the sequence of each plasmid by Sanger sequencing using the pX330 sequencing primer. We will call this plasmid pX330-sgRNA.
3.3 Transfection of HEK293 or IMCD3 Cells
A different transfection reagent is used depending on the cell line ( see Note 7 ). Choose Section 3.3.2, step 17, for transfection of HEK293 cells and Section 3.3.3, step 18, for transfection of IMCD3 cells. Regardless of transfection reagent, three transfection conditions will be used: (1) a co-transfection with pX330-sgRNA and pAcGFP1-C1 (“CRISPR” transfection); (2) a transfection with pAcGFP1-C1 (“GFP-only” transfection); (3) a mock transfection without plasmid DNA (“non-transfected” condition). The latter two conditions act as positive and negative controls of transfection, respectively.
3.3.1 Seeding of Cells for Transfection
The day before transfection, seed cells into three wells of a 6-well plate ( see Note 8 ). Plate 1 × 105to 3 × 105cells/well in 2 mL/well complete medium. Leave to achieve a desired confluence of 50–80%, appropriate for transfection.
3.3.2 Transfection of HEK293 Cells
Label three sterile microcentrifuge tubes, one for each transfection condition.
In each tube, mix 3µL with 97µL and incubate for 0h 5m 0s at Room temperature.
Add the appropriate plasmid DNA to each tube and mix. For the CRISPR transfection, add 500ng and 500ng. For the GFP-only transfection, add 1µg. No plasmid DNA is added for the non-transfected condition. Incubate for 0h 15m 0s at Room temperature.
To each well of plated cells, add 100µL in a dropwise manner.
Return cells to the incubator for 48h 0m 0s, prior to cell sorting (see Note 9).
3.3.3 Transfection of IMCD3 Cells
Label six sterile microcentrifuge tubes. For each transfection condition, there will be one tube for Lipofectamine and one tube for plasmid DNA.
In each of the three tubes, mix 8.5µLwith 150µL and incubate for 0h 5m 0s at Room temperature.
In the remaining three tubes, mix 150µL with the appropriate plasmid DNA. For the CRISPR transfection, add 1.25µg (obtained in step 3.2.4 ) and 1.25µg. For the GFP-only transfection, add 2.5µg. No plasmid DNA is added for the non-transfected condition.
Add each plasmid mixture to a separate tube of Lipofectamine and mix well. Incubate for 0h 30m 0s at Room temperature.
To each well of plated cells, add 300µL in a dropwise manner.
Return the cells to the incubator for 48h 0m 0s, prior to cell sorting ( see Note 9 ).
3.4 Group Sort by Fluorescence-Activated Cell Sorting (FACS)
Label a 15 mL centrifuge tube for use as a CRISPR sample collection tube. Add 500µL to the tube, put it On ice, and set aside.
Wash the CRISPR and non-transfected wells of cells twice each with 1mL/well sterile DPBS ( see Note 10 ).
Add1mL/well trypsin-EDTA and incubate at 37°C until cells dissociate.
Add 2mL/well complete media and transfer each 3mL to a 15 mL centrifuge tube.
Pellet cells by centrifugation at 200x g.
Discard the supernatant and resuspend cells in 300µL ( see Note 11 ).
Pass cells through a 70 μm cell strainer to remove clumps ( see Note 12 ).
Put the tubes directly On ice.
Proceed directly to FACS ( see Note 13 ). Use non-transfected cells to discern single cells from cell clumps when setting up the cell sorter. Isolate GFP-positive cells from the CRISPR sample and collect these in the collection tube (prepared in Section 3.4, step 19 ). Run the entirety of the sample or collect at least 5 × 105 cells per sample ( see Note 14 ). Keep collected cells On ice.
In a sterile safety cabinet, take one-half of the GFP-positive CRISPR sample and seed them in a 25 cm2 tissue culture-treated flask containing 5mL. Return the cells to the incubator and continue to grow them for 336h 0m 0s, passaging as and when necessary ( see Note 15 ).
Separately pellet the remaining GFP-positive CRISPR cells and the non-transfected cells for gDNA extraction, by centrifugation at 300x g. Discard the supernatant. Proceed directly to the next step, or store the cell pellets at -20°C.
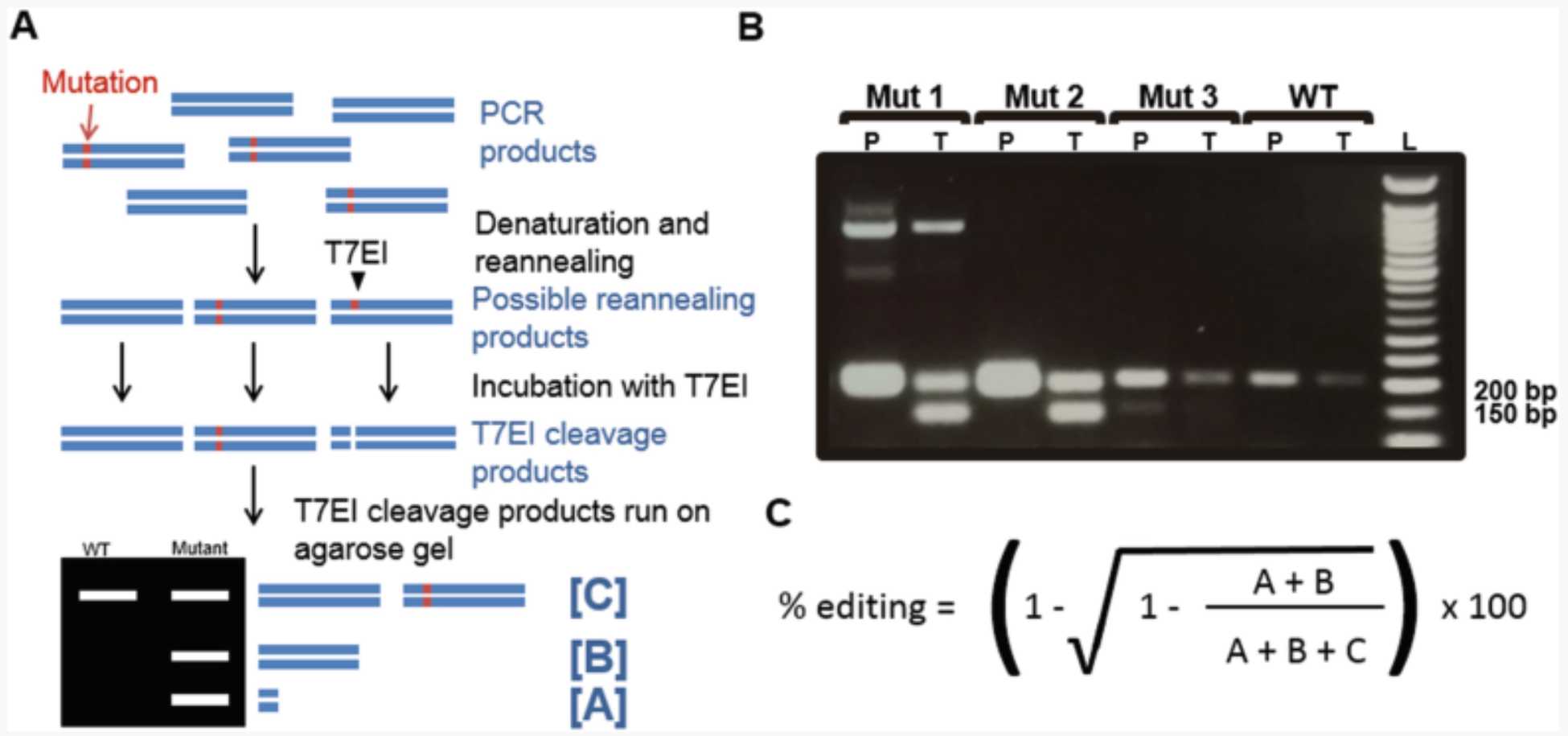
3.5 T7 Endonuclease I (T7EI) Assay
Extract genomic DNA (gDNA) with the DNeasy Blood and Tissue kit, following the manufacturer’s instructions.
Use the extracted gDNA as template to perform a 50µL PCR reaction using the genotyping primers (designed in Section 3.1, step 10 ) and Pwo Master ( see Note 16 ).
Run 25µL of the PCR in an agarose gel (adjusting the agarose percentage depending on the expected size of the PCR product) against a 1 kb DNA ladder to check whether there is a single product ( see Note 17 ).
In a 0.2 mL PCR tube, prepare a 19µL reaction containing 10µL PCR reaction, 2µL , and 7µL.
Melt the PCR products in a thermocycler using the following conditions: 95°C for 0h 5m 0s , ramp down to 85°C at −2 °C/s, ramp down to 25°C at −0.1 °C/s, and hold at 4°C.
Add 1µL to the melted PCR products and incubate at 37°C for 1h 0m 0s.
Run the T7 cleavage products in an agarose gel and look for cleavage of the PCR product ( see Notes 17 and Note 18 ; Fig. 3).
3.6 Single-Cell Sorting of CRISPR Cells to Establish Clonal Cell Lines
3.6.1 Preparation of Conditioned Media
In a 50 mL centrifuge tube, make up 16mL per 96-well plate ( see Note 19 ), by mixing 8mL with 8mL medium removed from a 75 cm2flask of cells at ~70% confluence ( see Note 20 ).
Sterile-filter the conditioned medium with a 0.2 μm filter ( see Note 21 ).
Fill a 96-well plate with 150 μL/well. Place the 96-well plate in the 37°C incubator.
3.6.2 Preparation of CRISPR Cells for FACS
Wash cells in 25 cm2 flask twice with 5mL .
Wash cells in 25 cm2 flask twice with 5mL .
Add 4mL and transfer cell suspension to a 15 mL centrifuge tube.
Pellet cells by centrifugation at 200x g.
Discard the supernatant and resuspend cells in 300µL( see Note 11 ).
Pass cells through a 70 μm cell strainer to remove clumps ( see Note 12 2 ).
Put the tubes directly On ice.
Proceed directly to FACS with the cell suspension and the 96-well plate containing conditioned media. Sort a single cell into each well of the 96-well plate ( see Note 22 ).
Spin the 96-well plate at 200x g and immediately return them to the incubator ( see Note 23 ).
3.6.3 Expansion of Cell Colonies
Incubate the cells for 2 weeks (336h 0m 0s), to allow colonies to establish ( see Note 24 ).
After 2 weeks, check individual wells under the microscope to identify those that contain a cell colony. Monitor cell confluence in these wells over the ensuing days.
When a well is confluent ( see Note 25 ), wash the cells twice in 200µL . Then dissociate cells in 30µL and transfer them directly into 1mL in a well of a 12-well plate ( see Note 26 ). Return the cells to the incubator and monitor their growth over the following days.
When a well of a 12-well plate is confluent, wash the cells twice with 1mL .Then dissociate cells in 150µL and transfer directly to a 25 cm2 flask containing 5mL. Return the cells to the incubator and monitor their growth over the following days.
When a 25 cm2 flask is confluent, passage the cells into two new 25 cm2 flasks. Return these flasks to the incubator.
When the two 25 cm2 flasks have reached ~70% confluence, freeze down and store the cells from one of the flasks.
With the remaining flask, wash cells twice with 5mL, and then dissociate in 1mL. Add 4mL and transfer the 5mL to a 15 mL centrifuge tube. Pellet the cells for 300x g and discard the supernatant. Proceed directly to the next step, or store the cell pellet at -20°C.
3.7 Genotyping of Clones
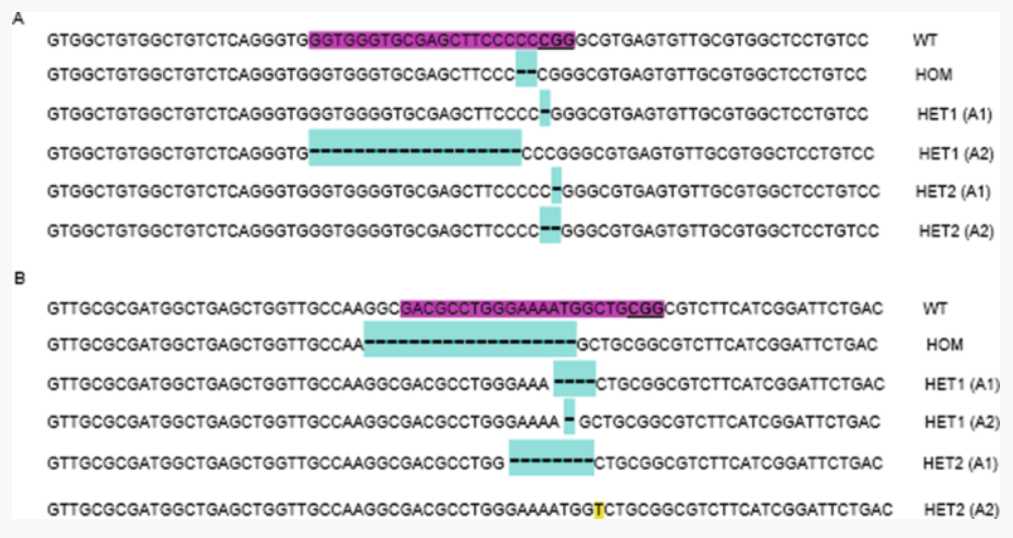
This method describes the genotyping of a single-cell colony.
Extract genomic (g) DNA with the DNeasy Blood and Tissue kit, following the manufacturer’s instructions.
Use the extracted gDNA as template to perform a 50µL PCR reaction with the genotyping primers (designed in Section 3.1, step 10 ).
Run the 50µL PCR in an agarose gel against a 1 kb DNA ladder. If there are multiple amplicons present, ensure that they are well resolved ( see Notes 27 and 28 ).
Gel-purify the PCR products.
Sanger sequence the extracted amplicon(s) to identify the presence and nature of mutations generated by CRISPR ( see Note 29 ; Fig. 4).