Generating CRISPR-Cas9-Mediated Null Mutations and Screening Targeting Efficiency in Human Pluripotent Stem Cells
Gregorio Alanis-Lobato, Gregorio Alanis-Lobato, Oliver J. Bower, Oliver J. Bower, Afshan McCarthy, Afshan McCarthy, Rebecca A. Lea, Rebecca A. Lea, Jasmin Zohren, Jasmin Zohren, Claudia Gerri, Claudia Gerri, James M.A. Turner, James M.A. Turner, Kathy K. Niakan, Kathy K. Niakan
Abstract
CRISPR-Cas9 mutagenesis facilitates the investigation of gene function in a number of developmental and cellular contexts. Human pluripotent stem cells (hPSCs), either embryonic or induced, are a tractable cellular model to investigate molecular mechanisms involved in early human development and cell fate decisions. hPSCs also have broad potential in regenerative medicine to model, investigate, and ameliorate diseases. Here, we provide an optimized protocol for efficient CRISPR-Cas9 genome editing of hPSCs to investigate the functional role of genes by engineering null mutations. We emphasize the importance of screening single guide RNAs (sgRNAs) to identify those with high targeting efficiency for generation of clonally derived null mutant hPSC lines. We provide important considerations for targeting genes that may have a role in hPSC maintenance. We also present methods to evaluate the on-target mutation spectrum and unintended karyotypic changes. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Selecting and ligating sgRNAs into expression plasmids
Basic Protocol 2 : Validation of sgRNA via in vitro transcription and cleavage assay
Basic Protocol 3 : Nucleofection of primed human embryonic stem cells
Basic Protocol 4 : MiSeq analysis of indel mutations
Basic Protocol 5 : Single cell cloning of targeted hPSCs
Basic Protocol 6 : Karyotyping of targeted hPSCs
INTRODUCTION
Human pluripotent stem cells (hPSCs) are established either from early embryonic epiblast progenitor cells of pre-implantation-stage embryos (Thomson et al., 1998) or following reprogramming of fibroblasts to generate induced pluripotent stem cells (Takahashi et al., 2007). hPSCs have the ability to differentiate into cells of all three germ layers and exhibit unlimited self-renewal (Nichols & Smith, 2009).
Appropriate spatial and temporal expression of genes is central to the regulation of pluripotency and early cell fate decisions (Martello & Smith, 2014; Ng & Surani, 2011). While the function of various distinct pluripotency-associated factors has been well explored in mouse embryonic stem cells (mESCs) (Niwa, 2007) and mouse embryos, the precise functional roles of factors in hPSCs and human embryos remain unknown and necessitate further investigation. The generation of loss-of-function mutations and subsequent phenotyping of the resultant null mutant cells is an informative approach to understanding the role of genes in these processes. While knockdown methods using siRNAs can provide some insight into gene function, it has been demonstrated that, due to a range of cell compensatory mechanisms, gene knockdowns do not necessarily phenocopy full knockouts (Rossi et al., 2015).
CRISPR-Cas9-mediated genome editing (Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013) is a relatively precise, efficient, easy-to-adapt and inexpensive method to generate and assess null mutations. By applying CRISPR-Cas9-mediated mutagenesis, the functional role of a gene in the maintenance of hPSCs can be assessed. Given the potential of hPSCs to differentiate into a variety of cell types (Cohen & Melton, 2011; Murry & Keller, 2008), targeting genes in hPSCs also allows for the evaluation of the functional role of factors in early lineage differentiation. Furthermore, a variety of methods to produce in vitro models of post-implantation human development generated from hPSCs have recently been developed (Shahbazi et al., 2017; Warmflash, Sorre, Etoc, Siggia, & Brivanlou, 2014), including the formation of multicellular mouse and human PSC aggregates called blastoids (Liu et al., 2021; Rivron et al., 2018; Yanagida et al., 2021; Yu et al., 2021) and human PSC−derived gastruloids (Moris et al., 2020). These more complex systems provide informative in vitro models to investigate putative developmental regulators. By aggregating hPSCs with null mutations in genes of interest to form these complex structures, their functional requirements can be assessed in a tractable model of early human development. hPSCs and their resulting structures are also a cellular context that is highly informative for refining CRISPR-Cas9 editing techniques before testing the role of a gene directly in precious human embryos. This approach, therefore, minimizes the number of embryos used in research and maximizes the knowledge that can be gained from these studies (Fogarty et al., 2017).
In addition, CRISPR-Cas9 genome editing can be used to generate and correct human disease models (Avior et al. 2016). CRISPR-Cas9-edited hPSCs can subsequently be differentiated into a variety of distinct cell types, and the resultant cells subjected to high-throughput drug screens and phenotypic analyses to inform on the nature of genetic diseases and to identify chemical compounds that may ameliorate disease (Elitt, Barbar, & Tesar, 2018). Moreover, a number of studies have been able to correct disease-associated mutations in patient-derived induced PSCs using homology-directed repair (Firth et al., 2015; Huang et al., 2015; Jacków et al., 2019), which relates to the possible use of this approach for future genetic therapies. Overall, the ability of hPSCs to differentiate into all somatic cell types allows for the opportunity to explore and modify a range of disease-causing mutant alleles that result in disorders of development, and to understand and correct disease phenotypes.
This article will serve to aid researchers in the generation of robust and well characterized null mutants in hPSCs. This document covers: the process of selecting optimal single guide RNAs (sgRNAs) using in silico prediction tools, followed by the process of ligation into an expression plasmid (Basic Protocol 1); the use of in vitro transcription and cleavage assays to screen the cutting efficiency of multiple sgRNAs (Basic Protocol 2); nucleofection of the sgRNA/Cas9 expression plasmid into primed hPSCs (Basic Protocol 3); using MiSeq Next Generation Sequencing to assess the editing efficiency of each sgRNA (Basic Protocol 4); the process of deriving single cell clones from targeted cells (Basic Protocol 5); and cytogenetic analysis of targeted cells (Basic Protocol 6).
STRATEGIC PLANNING
While the process of CRISPR-Cas9 targeting is simple and can be done relatively quickly, engineering a null mutation for a target gene of interest while minimizing off-targets requires a great deal of planning and consideration.
Although all genes are suitable for CRISPR-Cas9 targeting, a number of factors can enable more precise editing and a better chance of generating a null mutation. Well characterized target genes are beneficial, especially when functional domains of the associated protein are reported. To engineer null mutations to investigate gene function, it can be especially useful to direct the CRISPR-Cas9 genome editing strategy to target functional domains.
Some genes have undergone a large number of duplication events, which can lead to the formation of pseudogenes either on the same or different chromosomes in the genome relative to the target gene. The presence of multiple pseudogenes for a target gene of interest can make successful editing of the intended target more challenging if there is significant sequence homology. This is because off-target editing at the pseudogenes will result in a decrease in efficiency of the CRISPR-Cas9-mediated genome editing at the on-target site, and may lead to potential unintended off-target effects. Genes that are members of large gene families can also bring about similar difficulties in targeting, as some paralogs retain a high degree of homology. In some cases, however, this may be desired, as it could enable the simultaneous targeting of a whole gene family in order to investigate function. Overall, the ideal targeting strategy is to use a gene that is well understood, with unique sequence regions at or close to a known functional domain, avoiding the targeting of pseudogenes or any other region of the genome with sequence homology to minimize off-target effects.
The efficiency of CRISPR-Cas9-mediated genome editing can also be affected by the genomic variation that exists within the human population. CRISPR-Cas9 depends on a 20-base-pair (bp) guide RNA sequence to direct the Streptococcus pyogenes Cas9 (SpCas9) endonuclease to the target site to cleave double-stranded DNA. An important consideration in targeting human clinical samples, primary human cells, or human cell lines is the presence of single nucleotide polymorphisms (SNPs; Fig. 1A), or structural variants, between individuals at the CRISPR-Cas9 on-target site. Tools including the 1000 Genomes Project (1000 Genomes Project Consortium, 2015; Fig. 1B and 1C), as well as a number of statistical frameworks (Li, 2011), can prove useful for SNP calling and need to be utilized in the sgRNA and PCR primer design.
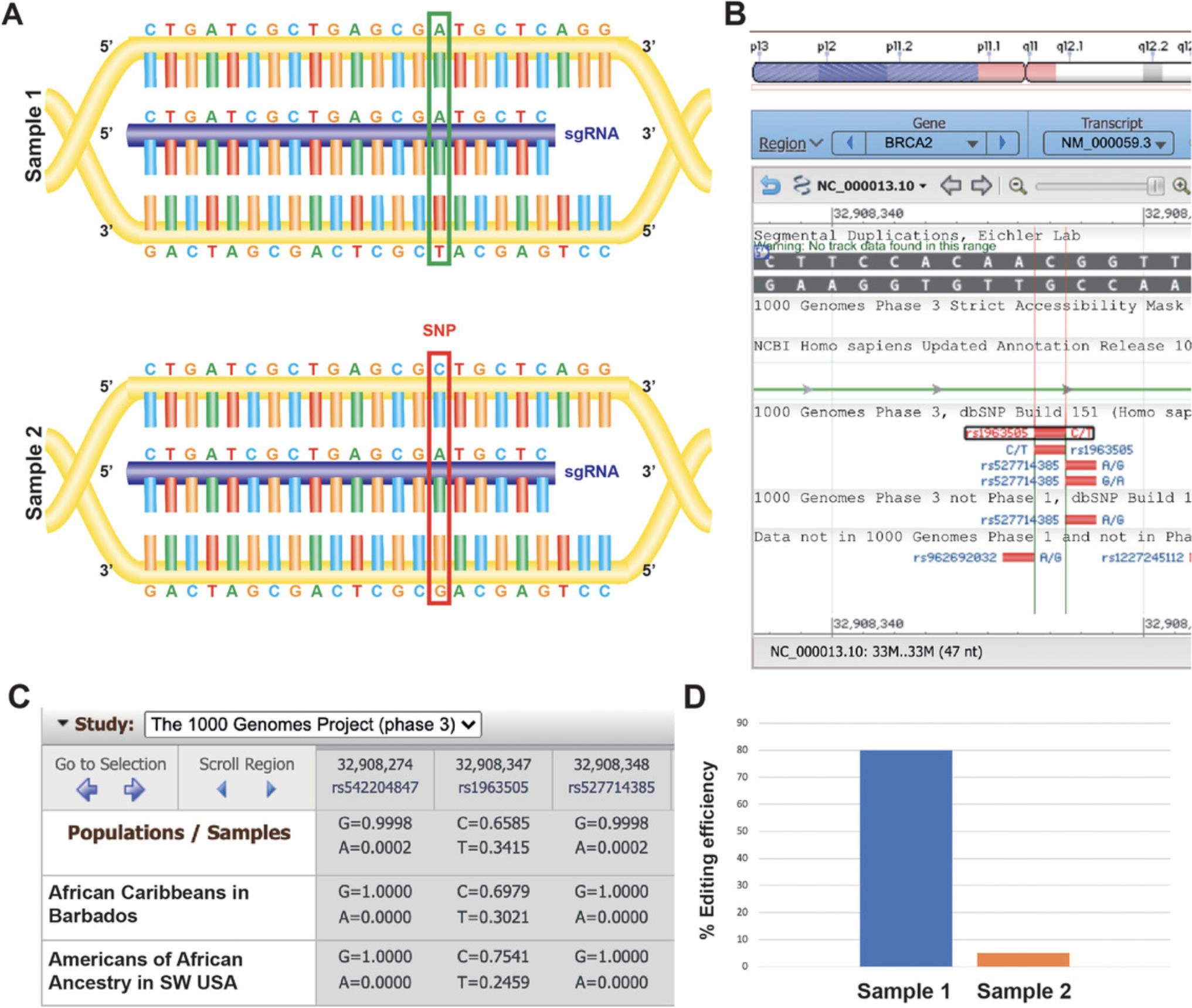
Similarly, the selection of primers used to amplify the targeted genomic DNA following CRISPR-Cas9 genome editing needs to be carefully considered and should be verified. PCR reactions will be crucial in the generation of amplicons for in vitro cleavage and in MiSeq analysis of insertion-deletion (indel) mutations. While gel electrophoresis informs the size of amplicons generated, it does not guarantee that the expanded region is necessarily the target region of interest. This is an important consideration especially when investigating genes within larger gene families or genes with one or more pseudogenes, as mentioned above. It is important to validate the sequence of all PCR amplicons to ensure that they precisely match the target region of interest. This will inform the identification of a guide RNA that has a higher likelihood of facilitating on-target null mutations following error-prone repair of DNA double-strand breaks.
Finally, verifying off-targets may be of paramount importance to some applications of CRISPR-Cas9 targeting, for example, in generation of disease models where the phenotype should be attributed only to on-target editing. To do this, it is important to make note of all potential off-target sites with ≤3 base-pair mismatches to the sgRNA. These sites should then undergo sequencing to assess for off-target editing. Basic Protocol 4 outlines the application of MiSeq to assess for indels at specific sequences. Alternatively, we also recommend Digenome-seq (Kim et al., 2015; Kim, Kim, Kim, Park, & Kim, 2016), CIRCLE-seq (Tsai et al., 2017), and GUIDE-seq (Tsai et al., 2015) for more comprehensive genome-wide assessments of possible off-target mutations.
Basic Protocol 1: SELECTING AND LIGATING sgRNAs INTO EXPRESSION PLASMIDS
sgRNA design
CRISPR-Cas9 gene targeting makes use of a 20-nucleotide-long variable CRISPR RNA spacer sequence (crRNA) complementary to the target sequence of interest, linked to a trans -activating CRISPR RNA (tracrRNA), which can together comprise an sgRNA. This sgRNA binds to a complementary 20-nucleotide-long region of interest located immediately upstream of a protospacer adjacent motif (PAM) sequence (NGG for SpCas9; Fig. 2A). The PAM sequence is not part of, nor complementary to, the sgRNA, but is necessary for Cas9 endonuclease to recognize the target site (Jinek et al., 2012). Cas9 will cleave DNA between 3 and 4 nucleotides upstream of the PAM sequence (Wu, Kriz, & Sharp, 2014). sgRNAs can target either the sense or antisense strand, as long as there is an NGG PAM sequence 3′ of the sgRNA sequence. SpCas9 can occasionally target NAG PAM sites; however, this requires an excess of Cas9/sgRNA (Wu et al., 2014).
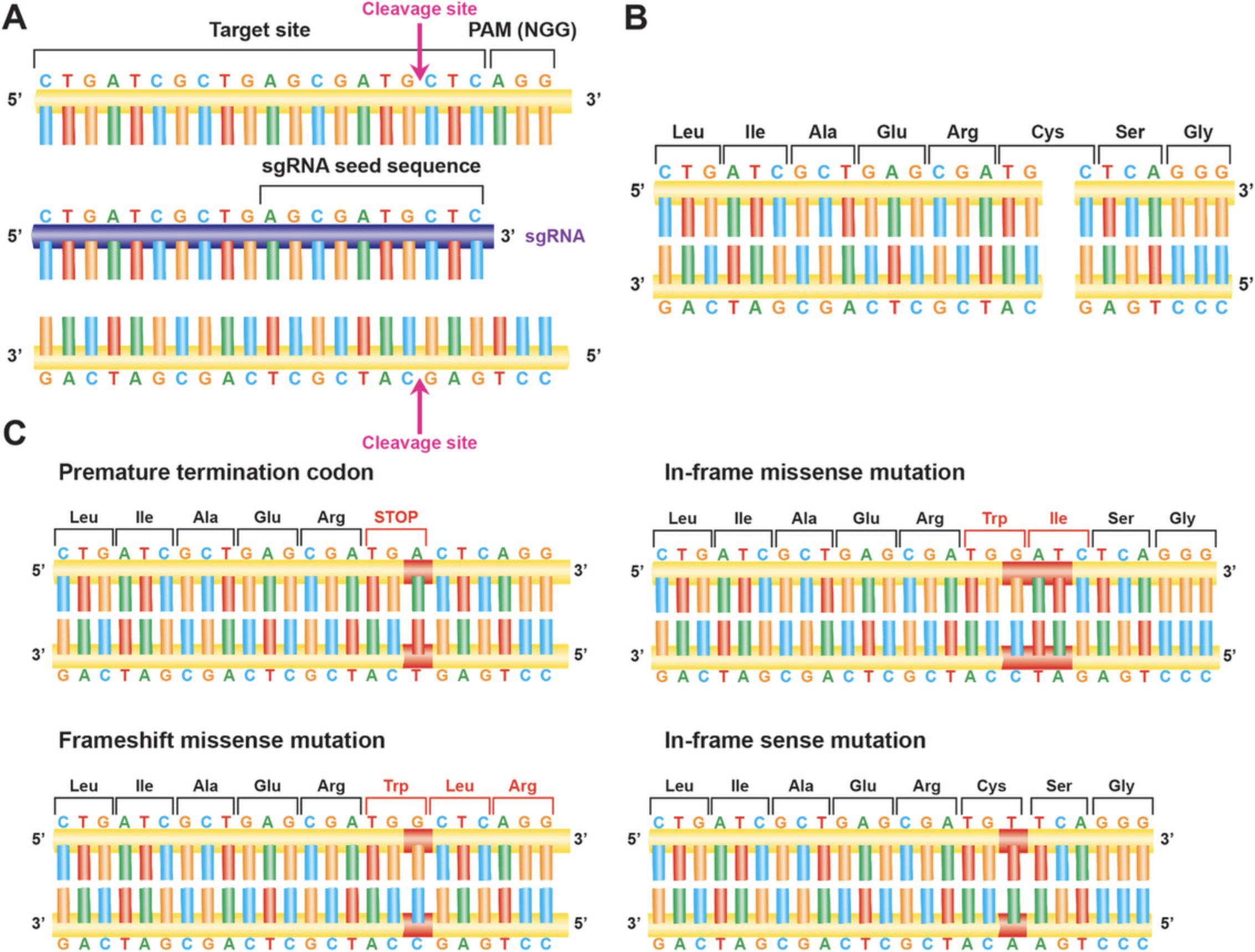
When eukaryotic cells detect double-stranded DNA breaks (DSBs), such as those produced by Cas9 activity, they initiate programs to repair them. One method comprises homology directed repair (HDR) via homologous recombination (Hsu et al. 2014). HDR involves strand invasion by a highly similar DNA followed by replication using this similar DNA sequence as a template (Capecchi, 1989). HDR therefore permits highly faithful replication of the original sequence. Another, more common method involves non-homologous end joining (NHEJ; Hsu et al., 2014). NHEJ involves resectioning and ligation of the two cleaved DNA ends, an error-prone process that leads to insertion and deletion of bases at the cleavage site (indels) (Fig. 2C). Finally, microhomology-mediated end joining (MMEJ) aligns short, microhomologous sequences from each side of the DSB. As MMEJ lacks a full-length repair template, it is a highly error-prone process, with the potential for larger indels surrounding the DSB compared to NHEJ (Sfeir & Symington, 2015; Seol et al., 2018; Zuccaro et al., 2020).
Successful gene targeting with CRISPR-Cas9 requires optimal sgRNA design. Appropriate sgRNAs should have high on-target efficiency and low off-target specificity; they should be well positioned around the target site of a gene in order to maximize rates of editing at the desired position. Loss-of-function null mutations can be generated when indel mutations lead to an out-of-frame mutation, disrupting the reading frame of protein translation, resulting in either a premature termination codon (PTC) or missense mutations at the target sequence. These mutations then cause abrogation of the resultant protein's functionality. Some algorithms exist to predict the propensity of sgRNAs for inducing frameshift mutations, though in silico predictions should always be tested systematically in the target cell type of interest because chromatin accessibility can vary between cell types, and this in turn can impact on CRISPR-Cas9 genome editing efficiency (Doench et al., 2016).
There are several approaches for identifying an appropriate target site for generation of a null mutant. In one approach, sgRNAs are designed to target near the 5′ end of the coding region. In this way, frameshift mutations can either result in the introduction of PTCs early in the gene sequence, or alternatively the whole of the coding region can be subjected to missense mutations. In the former, this should result in a highly truncated protein, while in the latter this might result in the production of a completely null mutant protein and degradation.
There are two important caveats to this technique. The first is that targeting too far upstream may result in generation and use of an alternative start codon for a known isoform of the gene, or cryptic splicing, allowing for translation of a protein with only a slightly truncated N-terminus. This resultant protein might, therefore, maintain some wild-type biological function or result in unexpected alterations in function (Biasio et al., 2007). The second is that production of a missense polypeptide might have unintended consequences on the cell; for example, it could trigger the unfolded protein response and result in cellular stress (Ma, Brewer, Alan Diehl, & Hendershot, 2002; Malhotra & Kaufman, 2007).
Alternatively, a key protein structural and/or functional domain can be targeted. An advantage to this approach is that a broader range of mutations may disrupt protein function and lead to a null mutant phenotype. As above, a frameshift mutation may lead to a PTC that could in turn trigger nonsense-mediated decay or lead to a truncated protein. However, the generation of in-frame missense mutations within a functional domain may also abolish gene function if they disrupt one or more key amino acids. Another approach is to target downstream of a functional domain. However, targeting too far downstream in the coding region may result in a substantially translated protein that may not have any loss of function, or may have unintended effects, for example leading to a dominant negative protein.
When aiming to efficiently generate null mutations, it may be beneficial to design sgRNAs with these approaches in mind and to determine which targeting approach to use by empirical testing, using the method outlined below.
Basic Protocol 2: VALIDATION OF sgRNA VIA IN VITRO TRANSCRIPTION AND CLEAVAGE ASSAY
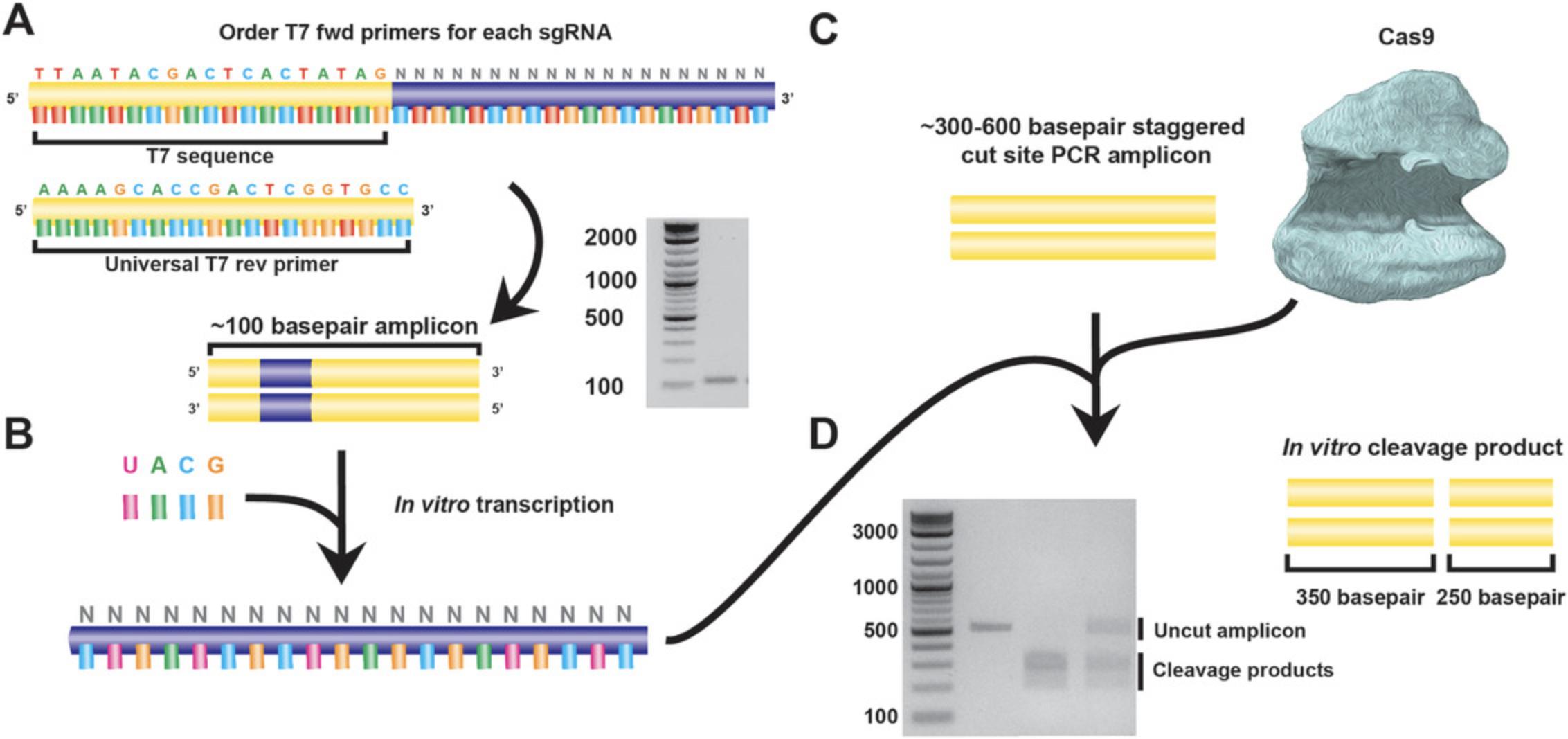
If many sgRNAs have been generated, it is preferable to narrow down potential guides using a relatively inexpensive and less laborious method such as an in vitro cleavage assay prior to directly targeting hPSCs.
After introduction into the pX459 plasmid (Fig. 3C), it is necessary to verify that the sgRNA can cleave genomic DNA in vitro before proceeding into live cells. To do this, in vitro transcription (IVT) and cleavage (IVC) assays are used. To perform IVT, primers are used to amplify the full-length sgRNA. One of the primers contains the T7 promoter sequence, which is used to perform in vitro transcription in the next step (Fig. 4B). Meanwhile, a PCR amplicon covering the sgRNA target locus is generated (Fig. 4C). The transcribed sgRNA and the genomic PCR amplicon are mixed together in the presence of a recombinant Cas9 protein, and the cleavage reaction is analyzed by agarose gel electrophoresis alongside a PCR amplicon control (Fig. 4D). In comparison to the single, uncleaved DNA band of the control, the IVC reaction product should reveal two distinct, shorter bands. To facilitate this, the PCR amplicon should be designed so that the cut site is asymmetrically located and products are of sufficient size for the cleaved products to be readily detected on a gel. Where possible, genomic DNA from the hPSC line used for downstream targeting should be used as a test of efficiency and to empirically determine if any polymorphisms may need to be considered.
Materials
-
Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs, cat. no. M0493S)
-
100 mM dNTP mixture (ThermoFisher Scientific, cat. no. 10297118)
-
T7 oligonucleotides for sgRNA for specific gene of interest (see step 1, below)
-
Guide-containing plasmid (Basic Protocol 1)
-
QIAquick PCR purification kit (Qiagen, cat. no. 28106)
-
DEPC-treated nuclease free (NF) water (Ambion, cat. no. AM9906)
-
RNaseZAP, or similar RNase cleansing agent
-
MEGAshortscript T7 kit (Life Technologies, Ambion, cat. no. AM1354)
- Alternative in vitro transcription kit: TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific, cat. no. K0441)
-
RNeasy MinElute Cleanup Kit (Qiagen, cat. no. 74204)
- Alternative RNA Cleanup Kit: RNA Clean and Concentrator (Zymo Research, cat. no. R1018).
-
Q5 High Fidelity Master Mix (New England Biolabs, cat. no. M0492S), or other preferred PCR reaction kit
-
Genomic DNA: gDNA is generated from H9 hESC's using the DNeasy blood and tissue kit (Qiagen, cat. no. 69504) according to the manufacturer.s instructions, DNA quantification is done using a Nanodrop spectrophotometer, and this is generally in the region of 50-100 ng/µl
-
Recombinant Cas9 protein (Toolgen, cat.no TGEN_CP3; other commercial sources of the Cas9 protein are also available such as Alt-R S.p. HiFi Cas9 Nuclease V3, IDT, cat. no. 1081060)
-
10× NEBuffer 3 (New England Biolabs, cat. no. B7003S)
-
10× bovine serum albumin (BSA; New England Biolabs, cat. no. B9001S)
-
RNase A (Promega, cat. no. A797C)
-
RNase-free 1.5-ml microcentrifuge tubes (Ambion, cat. no. AM12400)
-
PCR thermal cycler
-
37° and 65°C water baths
-
Filter pipette tips
-
Nanodrop spectrophotometer
-
Additional reagents and equipment for agarose gel electrophoresis (see Current Protocols article: Voytas, 2001)
T7 amplification
1.Order T7 primers for each sgRNA.
- Fwd: 5′-TTAATACGACTCACTATAG NNNNNNNNNNNNNNNNNNNN-3′
where N × 20 is the sgRNA.
- Rev: 5′ -AAAAGCACCGACTCGGTGCC -3′
The reverse primer is universal to the pX459 plasmid and does not require an attached sgRNA.
2.Perform a PCR reaction to amplify out the T7+sgRNA sequence:
Reaction mixture:
| 5× Q5 reaction buffer | 20 μl |
| 10 nM dNTPs | 2 μl |
| 10 μM forward T7 primer | 5 μl |
| 10 μM reverse T7 primer | 5 μl |
| Guide-containing plasmid (Basic Protocol 1) | 1 μl |
| Q5 Hot Start DNA Polymerase | 1 μl |
| 5× Q5 Enhancer (from Q5 kit) | 20 μl |
| NF water | 46 μl |
Thermal cycling program:
| 1 cycle of: | 98°C for 30 s |
| 25 cycles of: | 98°C for 7 s |
| 60°C* for 15 s | |
| 72°C for 50 s | |
| 1 cycle of: | 72°C for 2 min |
| Hold at 4°C indefinitely |
3.Purify the PCR product using the QIAquick PCR purification kit, according to the manufacturer's instructions, with elution into 20 μl NF water.
4.Take 2 μl of the purified product and run on a 2% TAE agarose gel at 100 V for no more than 1 hr (see Current Protocols article: Voytas, 2001).
- The amplified region should be approximately 100 bp in length.
In vitro transcription
In the following steps, it is important to adhere to RNase-free procedure: wiping down of surfaces and gloves with a product like RNaseZAP, use of filter pipette tips, use of RNase-free tubes, and working in a timely and careful manner on ice in order to minimize RNase contamination and sample degradation.
5.Thaw the frozen MEGAshortscript T7 kit reagents on ice, except for the 10× T7 reaction buffer, which should be thawed at room temperature.
6.Prepare the following IVT reaction in RNase-free tubes. Do not make a master mix; pipette each reagent individually into the required number of IVT tubes, as per manufacturer's instructions.
| T7 10× reaction buffer | 2 μl |
| T7 ATP solution | 2 μl |
| T7 CTP solution | 2 μl |
| T7 GTP solution | 2 μl |
| T7 UTP solution | 2 μl |
| Purified T7 guide DNA (step 4) | 8 μl |
| T7 enzyme mix | 2 μl |
7.Gently flick tubes to mix, pulse spin to bring liquid to bottoms of tubes, and incubate at 37°C for 3 hr.
8.Add 1 μl of TURBO DNase (from kit) and incubate at 37°C for a further 15 min.
- This step is optional but recommended to eliminate residual DNA
9.Purify the sgRNA using either a QIAGEN RNAeasy MiniElute Cleanup Kit or Zymo RNA Clean and Concentrator kit.
-
Follow the manufacturer's instructions, with elution into 20-50 μl of NF water
-
Immediately take a 1-3 μl aliquot for quantification and gel electrophoresisExpected concentrations are variable and can be gene- and gene-region dependent, but generally a concentration of 150-2000 ng/μl can be achieved with a 260/280 nm ratio of >1.8 and a 260/230 nm ratio of >1.8.The 18s rRNA positive control concentration should be approximately 2000 ng/μl.
-
Quickly freeze the remaining RNA at −80°C.
10.Mix together 2 μl of the purified RNA with 2 μl of the MEGAshortscript T7 RNA loading buffer. Incubate at 95°C for 5 min.
11.Run the sgRNA/loading buffer on a 2% TBE agarose gel at 100 V for 40 min (see Current Protocols article: Voytas, 2001).
Amplicon production
12.Design PCR primers to amplify a product roughly 200-500 bp in length, with the expected CRISPR-Cas9 cut sites site located off-center (e.g., 300/sgRNA site/200 for a 500-bp amplicon).
13.Identify the optimal conditions for PCR amplification by performing reactions across an annealing temperature gradient. It may be necessary to adjust other parameters, such as the nature of the DNA polymerase, concentration of MgCl2, or chemistry of the buffers used. Once a clean amplicon is produced, sequence the amplicon to ensure that the PCR primers are specific and amplifying the gene of interest.
14.Perform a PCR reaction to generate 50 μl of the amplicon with the identified optimal conditions from step 13.
Reaction mix:
| 2× Q5 master mix | 25.0 μl |
| 10 μM forward amplicon primer | 3.0 μl |
| 10 μM reverse amplicon primer | 3.0 μl |
| Genomic DNA | 2.0 μl |
| NF water | Up to 50 μl |
Thermal cycling conditions:
| 1 cycle of: | 98°C for 5 min |
| 35 cycles of: | 98°C for 30 s |
|
55°-65°C for 30 s* |
|
| 72°C for 30 s | |
| 1 cycle of: | 72°C for 2 min |
| Hold at 4°C indefinitely |
15.Purify the amplicon product using the QIAquick PCR purification kit.
- Follow manufacturer's instructions
16.Take 2 μl of the purified amplicon product and run on a 2% TAE agarose gel at 100 V for 1 hr (see Current Protocols article: Voytas, 2001).
17.Quantify the purified amplicon product by Nanodrop spectrophotometry.
In vitro cleavage assay
18.Set up the following incubation reaction mixture in 1.5-ml RNase-free microcentrifuge tubes:
| Cas9 protein | 200 ng |
| sgRNA (step 9) | 100 ng |
| PCR product (target; step 15) | 100 ng |
| 10× NEBuffer 3 | 1 μl |
| 10× BSA | 1 μl |
| Nuclease-free water | To 10 μl |
Incubate at 37°C for 1 hr.
19.Add 4 μg of RNaseA and incubate for a further 15 min at 37°C.
20.Heat inactivate the reaction by incubating at 65°C for 5 min.
21.Analyze the results on a 2% TAE agarose gel at 100 V for 60 min (see Current Protocols article: Voytas, 2001), running a sample of uncut amplicon beside each IVC reaction mixture, to demonstrate the cutting ability of each sgRNA.
Basic Protocol 3: NUCLEOFECTION OF PRIMED HUMAN EMBRYONIC STEM CELLS
Following the in vitro test of cleavage efficiency, the next step is to perform nucleofection of hPSCs with the most promising sgRNA(s) (Fig. 5A). Testing in hPSCs allows for the analysis of indel efficiency and mutagenic spectrum via subsequent MiSeq analysis. This step is helpful for further refining sgRNA selection, as the nature of indels generated and their relative frequency is specific to each sgRNA. Overall, for a null mutant, it is preferable to select a guide that consistently generates a high proportion of indel mutations that lead to frameshift mutations.
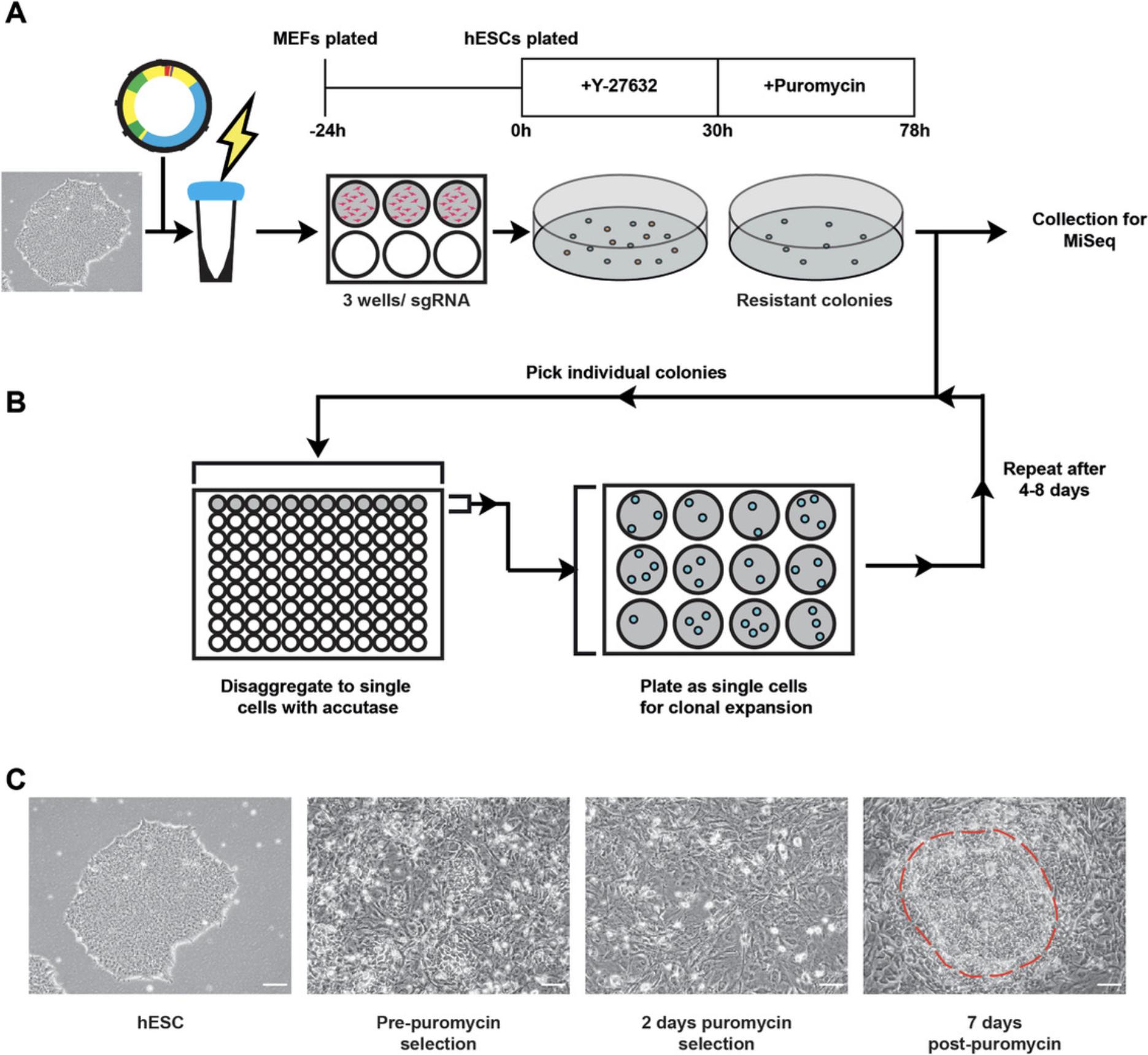
Mitotically inactivated mouse embryonic fibroblasts (MEFs) are plated the day before nucleofection and are used to aid the attachment and growth of the nucleofected hPSCs. This protocol uses MEFs at a density of 1 × 106 cells per well of a 6-well plate (9.5 cm2 growth area per well). This protocol also uses 2 × 106 cells of the H9 line of human embryonic stem cells per nucleofection of one sgRNA/condition. The 2 × 106 cells are then plated across 3 wells of a 6-well plate. It is likely that the number of cells required will need to be tested empirically, especially if performing the nucleofection in a cell line other than H9 hPSCs. This is due to inherent differences between hPSC lines and lab-specific cell culture techniques. It is good practice to use the GFP plasmid provided in the Lonza kit as a control for all nucleofection experiments. This will ensure that all reagents are working and that the procedure was followed accurately. Key parameters that should be tested empirically include method of disaggregating cells to single-cell density, number of cells placed per nucleocuvette, nucleofection program, concentration of plasmid added to the cuvette, and density of hPSC prior to nucleofection.
The culture medium of choice should be antibiotic free on the day of nucleofection, as antibiotics can decrease transfection efficiency. In the days after nucleofection, antibiotics can be used. After allowing cells to attach for 30 hr, the culture medium is then supplemented with puromycin for 48 hr in order to select for cells that have taken up the pX459 plasmid. Before nucleofecting cells, it is very important to perform a puromycin kill curve across a range of concentrations on the cell line that will be used, to ensure that the puromycin concentration eliminates all cells. This protocol uses mTeSR1 for culturing hPSCs, though alternative hPSC culture media, for example those that contain Knockdown Serum Replacement (KSR) together with fibroblast growth factor, or other media such as Essential 8 (E8), are likely to also be permissive for nucleofection.
Materials
-
DR4 MEFs, or other MEFs with neomycin and puromycin resistance
-
MEF medium (see recipe)
-
mTeSR1 (STEMCELL Technologies, cat. no. 85850), or hPSC culture medium of choice, without antibiotics
-
Y-27632 (ROCK inhibitor; Tocris, cat. no. 1254)
-
P3 Primary Cell 4D-Nucleofector X Kit L (Lonza, cat. no. V4XP-3012), or other nucleofector system
-
H9 line of human embryonic stem cells: Wicell, cat. no. WA09 (Thomson et al., 1998)
-
Dulbecco's phosphate-buffered saline (DBPS; ThermoFisher Scientific, cat. no. 14190-094)
-
pX459 plasmid with sgRNA at 4 μg/ml (see Basic Protocol 1)
-
Accumax (ThermoFisher Scientific, cat. no. 00-4666-56) or Accutase (STEMCELL Technologies, cat. no. 07920)
-
Puromycin (Sigma Aldrich, cat. no. P8833)
-
6-well plates
-
15- and 50-ml conical centrifuge tubes (Corning Falcon)
-
Automated cell counter or hemocytometer
-
4D-NucleofectorTM Core Unit (Lonza, cat. no. AAF-1002B), or other nucleofector system
-
4D-NucleofectorTM X Unit (Lonza, cat. no. AAF-1002X), or other nucleofector system
-
Centrifuge
-
5-ml stripette
-
Microscope
-
Additional reagents and equipment for basic cell culture techniques including counting cells (see Current Protocols article: Phelan & May, 2015)
Day before nucleofection
1.Warm MEF medium to 37°C for 15 min before using.
2.Plate MEFs in 3 wells of a 6-well plate per each condition per each sgRNA you intend to test.
3.Feed 50% confluent hPSCs with mTeSR1 supplemented with 10 μM Y-27632 16-20 hr prior to nucleofection. Return to incubator overnight.
Day of nucleofection
Have prepared a surplus of mTeSR1 without antibiotics and supplemented with 10 μM Y-27632 for diluting cell suspensions. Have sgRNA pX459 plasmids thawed and on ice. Have Accumax at room temperature, or Accutase pre-warmed to 37°C.
4.Make up P3 nucleofection reagent.
5.Retrieve plates from incubator. Aspirate mTeSR1/Y-27632.Wash each well with 1 ml DPBS then add 1 ml of Accumax or Accutase per well (of a 6-well plate). Ensure even coverage then return to incubator for 10-20 min.
6.Set up 4D nucleofector system using program CB150 (for H9 hESC), then input the number of cuvettes (i.e., the number of conditions) to nucleofect.
7.After incubation, quench the Accumax or Accutase with 1 ml mTeSR/Y-27632 per well, and tap to dissociate cells. Pipette gently 5-10 times with a P-1000 pipette tip to form a single-cell suspension, then collect into a 15-ml conical tube.
8.Take a 250-μl aliquot of the cell suspension for counting.
9.Count cells using either an automated cell counter or hemocytometer (see Current Protocols article: Phelan & May, 2015).
10.Calculate the number of cells in the cell suspension. Calculate the volume of the cell suspension needed for the necessary 2 × 106 cells per condition.
11.Pellet the required volume of cell suspension by centrifugation for 5 min at roughly 300-375 × g (1200 rpm), room temperature.
12.Prepare nucleocuvettes with 4 μg pX459 sgRNA or 4 μg GFP control (supplied in the Lonza nucleofection kit)
13.Prepare one 15-ml conical tube per condition with 6 ml of mTeSR1/10 μM Y-27632.
14.Resuspend cell pellet in the required amount of P3 nucleofection reagent (100 μl per condition).
15.Add 100 μl of cell suspension to each nucleocuvette. Tap the base gently against a work surface to ensure there are no air bubbles.
16.Initiate the 4D nucleofector system, running two samples at a time.
17.Using the supplied Pasteur pipettes, recover the cell suspension from each cuvette and add to the prepared 6 ml of mTeSR1/10 μM Y-27632 medium.
18.Label the MEF plates prepared in step 2 appropriately.
19.Aspirate MEF medium from plates. Using a 5-ml stripette, divide the 6 ml mTeSR1/10 μM Y-27632 cell suspension across 3 wells of a 6-well plate.
20.Check cells under the microscope, then shake side to side very gently and incubate for 30 hr.
Day after nucleofection
21.After incubation for ∼30 hr, check for GFP expression in GFP control condition.
22.Prepare the required volume of mTeSR1 supplemented with puromycin and feed cells 2 ml per well.
23.Refresh mTeSR1/puromycin medium 24 hr later.
24.After 48 hr of puromycin selection, check GFP control condition to ensure all cells are dead.
25.Immediately following selection, it is unlikely there will be any visible hPSC colonies. Continue to feed cells with mTeSR without puromycin each day for 2-10 days, after which colonies should begin to appear
26.Collect cells after a set number of days for downstream applications
Basic Protocol 4: MiSeq ANALYSIS OF INDEL MUTATIONS
A number of approaches can be used to assess the frequency and types of indels observed from CRISPR-Cas9 mutagenesis. While Sanger sequencing can be used to assess mutagenicity in other systems (Dehairs, Talebi, Cherifi, & Swinnen, 2016), it is not recommended for this protocol. The extraction of bulk populations of hPSCs will result in a very large array of indels and frequencies, which will make the resulting sequence difficult to interpret, even with the assistance of decomposition algorithms (Brinkman, Chen, Amendola, & van Steensel, 2014).
CRISPR-Cas9 mutagenesis, if performed with minimal off-targets, should result in a large amount of relatively small indel mutations at a specific site in the genome. The number of different permutations of mutations generated makes these sequences ideal for Next Generation Sequencing (NGS) approaches such as MiSeq. MiSeq analysis relies on PCR amplification of a 250-450 bp region flanking the target site using primers with MiSeq sequencing adapters at their 5′ and 3′ ends. This PCR reaction will, therefore, amplify all observed indel mutations in the targeted population. The PCR product undergoes a purification step with AMPure XP beads to remove contaminants left over from the PCR reaction, and is then submitted for sequencing. MiSeq analysis provides an in-depth understanding of the mutations produced, and therefore can be used to determine if an sgRNA is highly efficient at facilitating mutations, including the evaluation of the proportion of frameshift mutations produced.
Materials
-
Harvested nucleofection cell pellets (Basic Protocol 3)
-
QIAGEN DNeasy Blood and Tissue Kit (QIAGEN, cat. no. 69506)
-
Q5 High Fidelity Master Mix (New England Biolabs, cat. no. M0492S)
-
AMPure XP beads (Beckman Coulter, cat. no. A63881)
-
MiSeq PCR amplicon primers (see steps 1 and 2 below)
-
80% ethanol, freshly prepared
-
Reagent reservoir/trough
-
Multichannel pipettes
-
96-well PCR plates: the PCR reactions can be prepared in either thin-walled PCR tubes, 8- or 12-well PCR strips or 96-well PCR plates depending upon the number of samples and these are available from a number of suppliers such as Star Labs or ThermoFisher Scientific.
-
Magnetic stand
-
Bioinformatics packages:
- dada2 (R package)
- ggplot2 (R package)
- optparse (R package)
- RACER
- BWA
- CrispRVariants (R package)
- rtracklayer (R package)
- Biostrings (R package)
- seqinr (R package)
- GenomicFeatures (R package)
- glue (R package)
-
Additional reagents and equipment for agarose gel electrophoresis (see Current Protocols article: Voytas, 2001)
1.For MiSeq analysis, generate and order a set of PCR primer oligonucleotides surrounding the target site.
2.Once the optimal conditions and temperature for a set of primers have been identified, order the primers with the following MiSeq adapter sequences:
- Forward primer oligo: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-[Forward primer sequence]-3′
- Reverse primer oligo: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-[Reverse primer sequence]-3′
3.Extract DNA from each nucleofection sample from Basic Protocol 3 using the QIAGEN DNeasy Blood and Tissue Kit according to the manufacturer's instructions.
4.Prepare a 50 μl MiSeq primer PCR reaction for each sample and perform thermal cycling in a 96-well PCR plate or 8- or 12-well PCR strip, depending on the number of samples:
Reaction mix:
| 2× Master Mix | 25.0 μl |
| Forward primer | 3.0 μl |
| Reverse primer | 3.0 μl |
| Nuclease-free water | 17.0 μl |
| DNA from nucleofection sample | 2.0 μl |
Thermal cycling conditions:
| 1 cycle of: | 98°C for 30 s |
| 25 cycles of: | 98°C for 7 s |
|
55°-65°C for 30 s |
|
| 72°C for 30 s | |
| 1 cycle of: | 72°C for 2 min |
| Hold at 4°C indefinitely |
5.Bring the AMPure XP beads to room temperature 1 hr before proceeding.
6.Centrifuge the MiSeq primer PCR reaction plate for 1 min to collect any condensation from the lid.
7.Vortex the AMPure XP beads for approximately 1 min to ensure beads are evenly distributed. For each sample, pour an equal volume to the PCR reaction product of the AMPure XP beads across a trough (i.e., add 50 μl per sample to the reagent trough).
8.Using a multichannel pipette, add 50 μl of the AMPure XP beads to each sample in the PCR reaction plate.
9.Gently pipette the mixture up and down 10 times to distribute evenly.
10.Incubate at room temperature without shaking for 5 min.
11.Place the PCR reaction plate on top of a magnetic stand. Position the magnets so that the AMPure XP beads pellet on one side of the tubes, to allow pipetting without disturbing the pellet. Allow the beads to pellet for approximately 2 min.
12.Working carefully and with a multichannel pipette, remove and discard the supernatant from the PCR reaction plate tubes. If beads are drawn up by the pipette, return the volume and allow the beads to re-pellet on the side of the tube.
13.With the PCR reaction plate still on the stand, add 200 μl of freshly prepared 80% ethanol to each sample on the plate.
14.Incubate the PCR reaction plate on the magnetic stand for 30 s at room temperature without shaking.
15.Carefully remove and discard the supernatant.
16.Repeat steps 13 to 15 two further times.
17.Use a P-20 multichannel pipette to remove as much of the supernatant as possible.
18.With the PCR reaction plate still on the stand, allow the beads to air-dry for 10 min.
19.Remove the PCR reaction plate from the magnetic stand. Add 25 μl of 10 mM Tris, pH 8.5 (Buffer EB from the QIAGEN DNeasy kit) to each well.
20.Gently mix by pipetting up and down 10 times. Ensure that beads are fully resuspended.
21.Incubate at room temperature without shaking for 2 min.
22.Return the PCR reaction plate to the magnetic stand for 2 min to allow the beads to pellet.
23.Using a multichannel pipette, carefully transfer 20 μl of the supernatant to a set of labelled tubes to be sent for sequencing.
24.Add gel loading buffer to the remaining 5 μl of the supernatant and run this on a 1.5% TAE agarose gel (see Current Protocols article: Voytas, 2001) to verify that the PCR amplicon is present and of good quality.
25.Submit the purified 20 μl product for MiSeq.
MiSeq analysis
High-throughput amplicon sequencing, such as that generated with the MiSeq platform, provides a wealth of data regarding the permutations of mutations that exist within a heterogeneous population of targeted cells, such as a bulk pool of hPSCs following CRISPR-Cas9-mediated genome editing. This data output is in the format of .fastq files, which can be collapsed using a variety of tools to provide lists of individual sequences (variants) and the number of reads relating to each variant, that is, the frequency at which each variant was detected within the population. Tools include Galaxy (https://usegalaxy.org/; “Collapsed sequences” function) and the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/; “FASTQ/A Collapser” function). The FASTX-Toolkit can be used both online and via the command line. When dealing with a bulk population, the number of variants can reach many hundreds or even thousands, precluding manual analysis of the mutation spectrum.
Instead, bioinformatics tools, both online and command-line, can be used to automate the process of examining MiSeq sequence data (.fastq files) in order to assess the overall efficiency of CRISPR-Cas9-mediated mutation and the nature of the individual variants that arise. Aside from the sheer volume of sequence data generated from high-throughput sequencing, sequencing errors can also pose a problem for analysis of CRISPR-Cas9-mediated mutagenesis. For this reason, we utilize a bioinformatics pipeline for analysis of MiSeq data that incorporates quality control and unbiased correction of reads, prior to the analysis of mutation efficiency. This pipeline uses dada2 (Callahan et al., 2016) and RACER (Ilie & Molnar, 2013) to implement trimming and correction of MiSeq reads, followed by BWA (Li & Durbin, 2010) to map the corrected reads to a reference genome sequence. The mapped sequences are post-processed with samtools (Li et al., 2009), and then the analysis of indel formation efficiency is performed using the R package CrispRVariants (Lindsay et al., 2016). Further details about the purpose of each script and the variables to edit are found in the Statistical Analysis of the Commentary, below.
26.Perform quality control and clean-up of the MiSeq-generated .fastq files using the scripts available at https://github.com/galanisl/loh_scripts, within the folder ampliconSeq:
- First, execute the scriptcheck_fastq_quality.Rto check the quality of a small number of.fastqfiles.
check_fastq_quality.Ris intended to check, trim and filter around 12 reads at a time in an interactive manner.
-
Then, execute the scripttrim_and_filter.Rto correct all.fastqfiles in a given directory in a fixed manner.
-
Then, execute the scriptcorrect_fastq.sh.
27.With the trimmed and corrected read files (.fastq), perform the analysis of the rate of CRISPR-Cas9-mediated indel production using scripts from the github repository: https://github.com/jzohren/crispr-miseq:
-
First, create theinfo_file.csv.
-
Then, execute the scriptcrispr.sh.
Thecrispr.shscript will subsequently launch thecrisprvar.rscript.
28.Optimal guides are generally those that consistently produce frameshift mutations in the gene coding region.
Basic Protocol 5: SINGLE-CELL CLONING OF TARGETED hPSCs
After performing MiSeq analysis to identify the sgRNA sequence that facilitates the most efficient production of frameshift mutations, the next step is to derive a clonal population of the null mutant hPSCs by single-cell cloning. This step involves setting up a nucleofection as per Basic Protocol 3 (Fig. 5A), then picking the colonies that are generated to subsequently disaggregate into single cells. The process of picking colonies involves manual scoring and picking of colonies from the plate, followed by immersion in Accutase and subsequent plating into 12-well plates to expand (Fig. 5B). This process should normally be repeated to increase the likelihood of generating a clonal population that comprises cells with identical genotypes. This is because in the original plating some colonies could have merged, resulting in a heterogeneous colony comprising two separate genotypes.
Materials
-
Matrigel-coated plates (see recipe)
-
Accutase (STEMCELL Technologies, cat. no. 07920)
-
mTeSR1 (STEMCELL Technologies, cat. no. 85850), or hPSC culture medium of choice, without antibiotics
-
Y-27632 (ROCK inhibitor; STEMCELL Technologies, cat. no. 72302)
-
Nucleofected hPSCs (Basic Protocol 3; also see step 4, below)
-
Phosphate-buffered saline (PBS), without Ca2+ or Mg2+ (Dulbecco's PBS from ThermoFisher Scientific, cat. no. 14040141)
-
12-well plate
-
96-well plate
-
Inverted/dissecting microscope
-
Repeat pipettors and P-20 and P-200 tips
-
37°C biological incubator
1.Prepare 12 wells of a 12-well plate with Matrigel as described in Reagents and Solutions. Incubate at room temperature for at least 1 hr before use.
2.Prepare 12 wells of a 96-well plate with 25 μl of pre-warmed Accutase.
3.Supplement 15 ml of mTeSR with 10 μM Y-27632.
4.Remove nucleofected plate from incubator. Aspirate medium and wash with 2 ml PBS per well.
5.Aspirate PBS and add 1 ml of PBS.
6.Identify suitable colonies for passaging. Place the plate on an inverted/dissecting microscope inside a laminar flow hood.
7.Using a P-20 pipette set at 5 μl, score around the colony to dislodge it.
8.Gently take up the colony and elute into one of the Accutase-prepared wells of the 96-well plate.
9.Repeat steps 7 and 8 until 12 colonies have been taken.
- As mentioned, work in batches of 5-10 min at a time then rejuvenate cells with mTeSR.
10.Place the 96-well plate into a 37°C incubator for 5 min to dissociate the colony to single-cell density.
11.During the incubation, aspirate Matrigel from the 12-well plate. Rinse with 0.5 ml of PBS, then add 1 ml of mTesR supplemented with 10 μM Y-27632.
12.After incubation, remove the 96-well plate from the incubator.
13.Pipette each well up and down with a P-200 pipette set at 25 μl to dissociate colonies to single cells. Check under the microscope to ensure this.
14.Transfer 25 μl from a well of the 96-well plate into one well of the 12-well plate.
15.Repeat step 14 until all 12 colonies have been disaggregated and plated into separate wells.
16.Gently shake plate side to side to ensure even distribution of cells, then incubate at 37°C overnight.
17.After 16-24 hr from plating, change the medium to mTeSR without Y-27632.Feed daily with mTeSR.
18.Optionally, after 4-8 days when colonies are of sufficient size, repeat steps 7-16 to ensure that colonies are derived from single clones and comprise a single homogenous population.
Basic Protocol 6: KARYOTYPING OF TARGETED hPSCs
Before embarking on experiments with clonal hPSC lines generated following CRISPR-Cas9-mediated genome editing, it is important to assess the cells for any gross karyotypic abnormalities. PSCs have a shortened cell cycle compared to most other cell types due to a shortened G1 phase, which results in an increased rate of replicative stress (Ahuja et al., 2016). This replicative stress can then result in DSBs, facilitating chromosomal duplication and translocation events (Zeman & Cimprich, 2014). hPSCs are therefore prone to acquiring part- or whole-chromosome abnormalities during routine culture (Draper et al., 2004). These aneuploidies can provide cells with a growth advantage (International Stem Cell Initiative, 2011), and therefore drive further genome instability. Additionally, this genomic instability can be exacerbated during the process of CRISPR-Cas9-mediated DNA double-strand breakage and repair, and further by the process of single-cell amplification (Garitaonandia et al., 2015) and the use of Rho kinase (ROCK) inhibitors (Weissbein et al., 2019). Indeed, there have been reports of large deletions, and even loss of whole chromosome arms, in response to genome editing in various cellular contexts (Shin et al., 2017; Adikusuma et al., 2018; Kosicki, Tomberg, & Bradley, 2018; Cullot et al., 2019; Owens et al., 2019; Zuccaro et al., 2020; Przewrocka, Rowan, Rosenthal, Kanu, & Swanton, 2020). Therefore, analyzing genome-edited cells for any such genomic aberrations is important because this may interfere with the interpretation of results generated from downstream applications.
There are a number of techniques commonly used to assess the karyotype of hPSCs, including G(Giemsa)-banded karyotyping (McIntire et al., 2021) and array comparative genomic hybridization (CGH; Wu et al., 2008; Elliott, Elliott, & Kammesheidt, 2010). However, these conventional methods of karyotyping are often slow, labor-intensive, and comparatively expensive. Additionally, they generally provide results at only low genomic resolution, ranging from ∼25 kb for array CGH to 3-10 Mb for G-banding (Stephenson et al., 2010). When screening a large number of genome-edited hPSCs, these limitations can present a significant bottleneck and delay experimental progress.
We recommend assessing the karyotype of hPSCs following CRISPR-Cas9-mediated genome editing through low-pass whole genome sequencing (WGS; Fogarty et al., 2017). This method has the benefits of providing karyotype analysis with rapid turnaround and relatively low costs. 0.1× coverage of WGS is sufficient for robust karyotyping analysis, which depends on the assumption that, in a karyotypically normal sample (without copy number variations (CNVs)), each genomic locus should provide roughly equal read counts, dependent upon their sequence content. Following low-pass WGS, this analysis can be performed bioinformatically. In brief, reads from low-pass WGS are aligned to the latest build of the human reference genome (hg19 build) using BWA version 0.7.17 and copy-number profiles generated using the R package QDNAseq version 1.24.0, using bins of size 100 kb. Further details about the purpose of each script and the variables to edit are found in the “Statistical Analysis.”
Materials
-
DNeasy Blood and Tissue Kit (QIAGEN, cat. no. 69506)
-
Buffer containing Tris·HCl (pH 8.5) without EDTA
-
Illumina DNA Prep (Illumina, cat. no. 20018704)
-
Illumina MiSeq System (Illumina, cat. no. SY-410-1003)
-
Nanodrop spectrophotometer/fluorometer
-
Bioinformatics Packages required:
- BWA
- QDNAseq (R package)
Nanodrop spectrophotometer
1.Extract total genomic DNA from hPSCs using a QIAGEN DNeasy Blood and Tissue Kit, or other preferred method.
2.Elute purified DNA into buffer containing Tris·HCl (pH 8.5) without EDTA.
3.Measure the concentration and purity of extracted DNA using a Nanodrop spectrophotometer/fluorometer.
4.Genomic DNA should be at a minimum concentration of 5 ng/μl, and a total amount of at least 100 ng DNA is ideal.
5.Prepare genomic DNA for sequencing using Illumina DNA prep.
6.Sequence the prepared libraries on an Illumina MiSeq System.
7.Perform copy-number analysis of low-pass WGS data using the scripts available at https://github.com/galanisl/loh_scripts, within the folder lowpassWGS.
-
First, execute the scriptalign_lowpassWGS.sh.
-
Then, execute the scriptgenerate_CNP.R.
REAGENTS AND SOLUTIONS
Matrigel-coated plates
- 1.Thaw Matrigel bottle (growth factor reduced Matrigel; BD Biosciences, cat. no. 356231) overnight at 4°C.
- 2.The next day, store microcentrifuge and pipette tips at −20°C for 1 hr to cool. Matrigel readily polymerizes at room temperature.
- 3.Prepare 6-well aliquots of Matrigel on ice by adding 2 mg of Matrigel per microcentrifuge tubes.
- 4.Store 6-well aliquots at −20°C until needed.
- 5.When needed, thaw a 6-well aliquot at 4°C for 20 min.
- 6.Quickly add the 6-well aliquot to 6 ml of Advanced DMEM/F-12 (ThermoFisher Scientific, cat. no. 12634-010). Mix gently.
- 7.Add 1 ml of the Matrigel/DMEM-F12 mix per well of a 6-well plate.
- 8.Swirl gently to coat entire well. Avoid bubbles
-
Allow to set for 1 hr at room temperature before usage
If not using immediately, cover with parafilm and store at 4°C until needed. Cooled Matrigel plates last for 1 week at 4°C. Ensure plates are brought to room temperature for at least 1 hr before usage.
MEF medium
See Table 2. Store at 4°C when not using.
| Component | Supplier | Cat. no. | Stock concentration | Final concentration | Volume used |
|---|---|---|---|---|---|
| Advanced DMEM/F12 | Invitrogen | 12634010 | - | 88% | 440 ml |
| Fetal bovine serum | Bioserum | S1818 | - | - | 50 ml |
| Glutamax | ThermoFisher Scientific | 35050038 | 200 mM | 2 mM (1%) | 5 ml |
| β-mercaptoethanol | Sigma | M6250 | 14.3 M | 0.1 mM | 3.5 μl |
| Optional: Penicillin/streptomycin | ThermoFisher Scientific | 15140-122 | 100% | 1% | 5 ml |
COMMENTARY
Background Information
The development of the hPSC field, from initial derivation as hESCs (Thomson et al., 1998), through to the production of iPSCs (Takahashi et al., 2007) as well as the refinement of culture conditions (Ludwig & Thomson, 2007), has led to a rapid advancement in the potential applications of these cells.
Concurrently, the field of genome editing has undergone rapid advancement. Two years before hESCs were first derived, zinc finger nucleases (ZFNs), restriction enzymes generated by the fusion of zinc finger DNA binding domains with DNA cleavage domains, were generated (Kim, Cha, & Chandrasegaran, 1996). These ZFNs were subsequently used in Xenopus oocytes, resulting in DNA cleavage and homologous recombination (Bibikova et al. 2001). With this, it became possible to engineer the zinc finger components to target any region of genomic DNA.
Next came the development of transcription activator−like effector nucleases (TALENs), working in a similar way to ZFNs, albeit using a TAL effector DNA binding domain in place of a zinc finger (Boch et al., 2009). This means TALENs are able to bind DNA specifically at individual base pairs, while zinc fingers usually recognize groups of three base pairs. Overall, both of these methods rely on protein/DNA interactions as their target design, meaning each target region requires a separate protein to be designed. As well, ZFNs require linkages between zinc fingers to be generated, while TALE motifs require Golden Gate assemblage (Engler, Gruetzner, Kandzia, & Marillonnet, 2009) to be correctly linked.
In contrast, CRISPR-Cas9-mediated genome editing relies on a fixed S. pyogenes Cas9 protein, driven by a variable sgRNA (Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013), which makes the design and engineering less complicated for non-specialists. Moreover, there are a number of expression vectors with the Cas9 gene, along with resistance genes and a site for sgRNA ligation. These features make the CRISPR-Cas9 system quick to implement for a relatively low cost.
CRISPR-Cas9 has been widely applied to target a number of genes in hPSCs. For example, Lee et al. (2019) used CRISPR-Cas9 to interrogate the role of FOXA2 in pancreatic differentiation of hPSCs. By generation of FOXA2 -null hPSCs, it was demonstrated that FOXA2 was necessary for pancreatic differentiation. In particular, chromatin accessibility and enhancer priming were disrupted in FOXA2 -null mutant hPSCs, impairing their ability to establish the pancreatic lineage transcription program.
In addition, Wang et al. (2017) investigated the functional role of the p53 family in hESC differentiation. By generating Trp53 , Trp63 , and Trp73 triple knockout hESCs, researchers demonstrated that this gene family drives activation of Wnt3, while also facilitating TGF-β signaling, which triggers mesendodermal differentiation. Overall, this study and others allow the investigation of gene regulatory networks that drive specific lineage differentiation in hESCs. Recently, CRISPR-Cas9-mediated null mutations of POU5F1 , SOX2 , and NANOG in naïve hESCs have suggested a role for these genes in preventing trophoblast stem cell differentiation (Guo et al., 2021).
In all, this demonstrates the utility of genetic studies in hPSCs to study gene function. Generation of null mutations is a classical method to investigate the functional requirement of genes in distinct cellular and developmental contexts. This, in conjunction with the unique differentiation potential of hPSCs, allows for the study of early developmental programs in vitro. These insights will prove crucial in the translation of hPSCs to regenerative approaches in the coming years (Eguizabal et al., 2019).
Critical Parameters
Selection of sgRNAs
An understanding of the protein structure of the gene being targeted is advantageous for CRISPR-Cas9 genome editing. Knowing the functional domains and their locations can help to identify key parts of the protein, and therefore coding sequence that needs to be eliminated to disrupt protein activity. However, CRISPR-Cas9 can still be used to target genes when protein structure is not well understood, but guides should be located relatively close to the 5′ end of the gene so that the resulting indels cause frameshift, missense mutations of the entire gene sequence.
A range of in silico prediction tools should be used for sgRNA design because each tool makes use of different algorithms and scoring methods to rank sgRNAs. On-target efficiency scores vary widely between prediction tools. If on-target scores are going to be used, it is important to check the model system that was used to generate the algorithms for these scores. For example, Doench et al. (2016) assessed sgRNA efficiencies in a range of human cancer cell lines; therefore, prediction algorithms that use this data should be taken cautiously when being applied to hPSCs. Overall, the main criterion of interest for sgRNA design should always be minimizing the prevalence of off-targets, not in silico on-target efficiency scores, which can be inaccurate depending on the cellular context under investigation.
Minimizing off-target homology is the most important parameter. While on-target efficiency scores are derived by algorithms, the off-target likelihood is simply based on sequence homology of the sgRNA to other regions in the genome. The greater the number of mismatches an sgRNA has to other regions in the genome, the lower the likelihood it will bind there and induce CRISPR-Cas9-mediated cleavage. Because of this, sgRNAs with mismatches of zero or mismatches of one base outside of the sgRNA seed sequences should be disqualified, as there is a very high likelihood of binding to these off-target regions. sgRNAs with mismatches located within the seed sequence (i.e., the 10-12 bases immediately 5′ of the PAM sequence) are more acceptable than mismatches located outside of the seed sequence. This is because mismatches in the seed sequence are more detrimental to the ability of CRISPR-Cas9 to bind and cleave DNA (Jiang & Doudna, 2017).
As mentioned above, sgRNAs should target functional domains, or be located towards the 5′ to cause frameshifts further downstream. This is done to ensure generation of a null mutant phenotype by eliminating all biological activity of the resultant protein. Sites that fail to eliminate functional domains of the protein are likely to generate unexpected results, including dominant negative phenotypes. This is because, while certain structural elements of the protein may be lost, it may still be able to compete for binding sites with other endogenous factors.
hPSC culture : The process of nucleofecting hPSCs places cells under a large amount of stress. Because of this, hPSC colonies should be stable and healthy in morphology (Fig. 5C). Cells should have been stably passaged at least three times after thawing. Moreover, cells should be growing in tight, compacted colonies, with minimal numbers of differentiating cells on the periphery.
Cell ploidy should be regularly checked, as hPSC grown for multiple passages can acquire gain or loss of chromosomal regions (Draper et al., 2004). These changes can disrupt targeting efficiency and compromise the pluripotency and behavior of the cells.
Troubleshooting
Plasmid ligation results in no colonies returning a successfully ligated plasmid
When transforming the ligated plasmid into bacteria, always prepare a water control plate, which will allow you to predict the rate of plasmid uptake. Poor cloning efficiency could be due to poor-quality purification of the plasmid. Consider using an alternative kit or check the existing one for contamination of buffers.
T7 guide DNA amplicon has a weak band when run on a gel
A poor T7 DNA product is likely due to poor action of the T7 primers. In this case, run a temperature gradient to optimize the T7 PCR reaction. Ensuring the T7 DNA amplicon is of good quality is important for generating a good in vitro transcription product later in the protocol.
In vitro transcription product does not produce clear bands on a TBE gel
Ensure that an RNase-free environment is maintained through the use of fresh and filtered pipette tips, RNaseZAP, wearing a face mask, working on ice, and use of fresh high-grade RNase-free water (aliquoting this can be helpful). Always perform each round of IVT reactions with the provided 18s rRNA control. If the control shows evidence of degradation, then this will indicate poor RNA handling. If available, make use of a PCR workstation to minimize contamination.
Moreover, ensure RNase-free handling in the extraction of the RNA, storing aliquots in RNase-free tubes and diluting in RNase-free loading dyes (such as the ones provided in the Zymo RNA clean and concentrator kit). Consider using alternative RNA extraction kits. While our lab uses the Zymo RNA Clean and Concentrator, other labs have used the Qiagen RNA extraction kit, while others have reported success with the NEB T7 transcription kit. If all of these kits also present issues in extracting RNA, consider a phenol/chloroform RNA extraction instead. Run the TBE gel for a shorter amount of time. RNase in the TBE in the open air can cause the RNA to degrade. The gel can be run for as little as 20 min, which should allow for discrimination of a band at 100 bp in length
In vitro cleavage assay does not work
Check the quality of RNA produced in the in vitro transcription reaction, and troubleshoot as above. Sequence the PCR amplicon target to ensure that it corresponds to the gene of interest and is devoid of any SNPs that could disrupt sgRNA binding. While the protocol above advises using the Q5 PCR kit, some sequences that are particularly GC rich could benefit from PCR with the Roche GC-Rich kit (cat. no. 12140306001). Check that the cut amplicon lengths are staggered and that the cut site is not within 20 bp of the amplicon start. Increasing the concentration of Cas9 protein can also improve in vitro cleavage. Consider using a positive control of a known sgRNA with high in vitro cutting efficiency to verify that reagents and technique are not the cause.
Absence of hPSC colonies post-nucleofection
Allow at least 3-4 days for colonies to return, hPSCs are not easily visible among the MEFs in the first few (0-2) days after puromycin selection. If colonies are still absent after allowing for this time, it suggests that the cells did not survive the nucleofection process. Ensure that hPSC are being grown in such a way as to promote their growth and maintain their pluripotency, as per “Critical Parameters. Ensure that cells spend as little time in P3 nucleofection buffer as possible; the solution is cytotoxic. Therefore, work quickly and efficiently while the cells are in P3 nucleofection buffer and ensure that the Falcon tubes of mTeSR + Y-27632 are pre-prepared to rapidly quench the buffer.
Perform a puromycin kill curve to ensure that the puromycin concentration used is no more than the concentration needed to kill all wild-type cells. Going above this concentration can cause toxicity to resistant cells. Typically, puromycin concentrations effective in hPSCs range between 0.20 and 0.70 μg/ml.
Low hPSC editing efficiency
Plasmid concentrations of <2000 ng/μl will not yield effective editing. Otherwise, consider editing parameters involved in the nucleofection of the cells. Check manufacturers suggestions, but the primary parameters to consider adjusting and testing empirically include: confluency, the number of days hPSCs are cultured prior to nucleofection, density of the cells that are nucleofected, the nucleofection program, and the concentration of plasmid used. We highly advise using the supplied Lonza GFP plasmid when testing parameters. For MiSeq analysis, we suggest using an established sgRNA of known efficiencies in hPSC as a positive control.
Statistical Analysis
Basic Protocol 4–MiSeq analysis
Check_fastq_quality.R This script generates figures displaying the read quality profiles of up to 12 .fastq files generated by MiSeq at a time. This allows one to assess the quality across the length of individual reads and determine whether and where reads may need to be trimmed to ensure that only data of sufficient quality is used in downstream analysis. To execute the script, one needs to:
- 1.Define the file path to the directory containing the MiSeq .fastq files in line #10;
- 2.Define the desired file path for the output of quality-filtered .fastq files in line #11;
- 3.From line #35 onwards, define the positions for read trimming, the acceptable number of sequencing “errors,” the acceptable number undefined bases (N) and a minimum acceptable quality score. In the provided example, these parameters are set to:
- a. truncate the reads at position 150 – truncLen=c(150,150);
- b. trim the first five 5′ bases of the reads – trimLeft = 5;
- c. accept at most five sequencing errors – maxEE=c(5,5);
- d. filter out all undetermined bases (Ns) – maxN=0; and,
- e. truncate reads when any single base has a quality score of 2 or less – truncQ=2.
This script will yield a series of .fastq files that have been adjusted to meet the parameters above. These trimmed and filtered .fastq files serve as the input for the correct_fastq.sh script below.
trim_and_filter.R This script will trim and filter all .fastq files in the same directory as the script. Unlike check_fastq_quality.R, this will not output figures to display read quality profiles. The trimmed and filtered reads will then be output into a fastq_flt file directory. These trimmed and filtered .fastq files serve as the input for the correct_fastq.sh script below.
correct_fastq.sh This script uses the package RACER to correct any likely sequencing errors within the reads generated by MiSeq. To execute this script, one needs to:
- 1.Create a text file called samples.txt, which is a simple list of sample names to be corrected (i.e., the names of the trimmed and filtered .fastq files generated above), with one name per line;
- 2.Define the location of the .fastq files to be corrected in line #10;
- 3.Define the location of an output folder, in which the corrected .fastq files will be deposited, in line #11; and,
- 4.Define a “genome size” in lines #14 and #15, against which to compare the MiSeq reads: in the provided example, this is set to 25,000.
info_file.csv This table (comma-separated value format; Fig. 6A, 7) contains vital information that is fed into the CrispRVariants package. It can list all of the samples (.fastq files) within a given directory. The fields are as follows:
- sample – the identifier of each .fastq file, one per forward and reverse read pair;
- gene – the name of the gene or locus that has been targeted by CRISPR-Cas9;
- seq – the 20 bp DNA sequence corresponding to the sgRNA sequence that was applied in the given sample;
- chr – the chromosome on which the CRISPR-Cas9 target sequence lies. The chromosome nomenclature must match that of the reference genome being used. For example, ‘1’ will not work with a reference genome that uses ‘chr1’ to denote chromosome 1, ‘chr1’ must be used instead.
- start – the genomic coordinates corresponding to the 5′-most base of the CRISPR-Cas9 target sequence;
- end – the genomic coordinates corresponding to the 3′-most base of the CRISPR-Cas9 target sequence;
- strand – the DNA strand (coding (+) or non-coding (-)) in which the CRISPR-Cas9 target sequence is found;
- plus – the number of base pairs that should be analyzed upstream of the 20 bp target sequence; and,
- minus – the number of base pairs that should be analyzed downstream of the 20-bp target sequence. These latter two parameters do not need to be equal, for instance, if one expects indel mutations to predominantly stretch downstream of the CRISPR-Cas9 target site, the “minus” value can be greater than the “plus.”
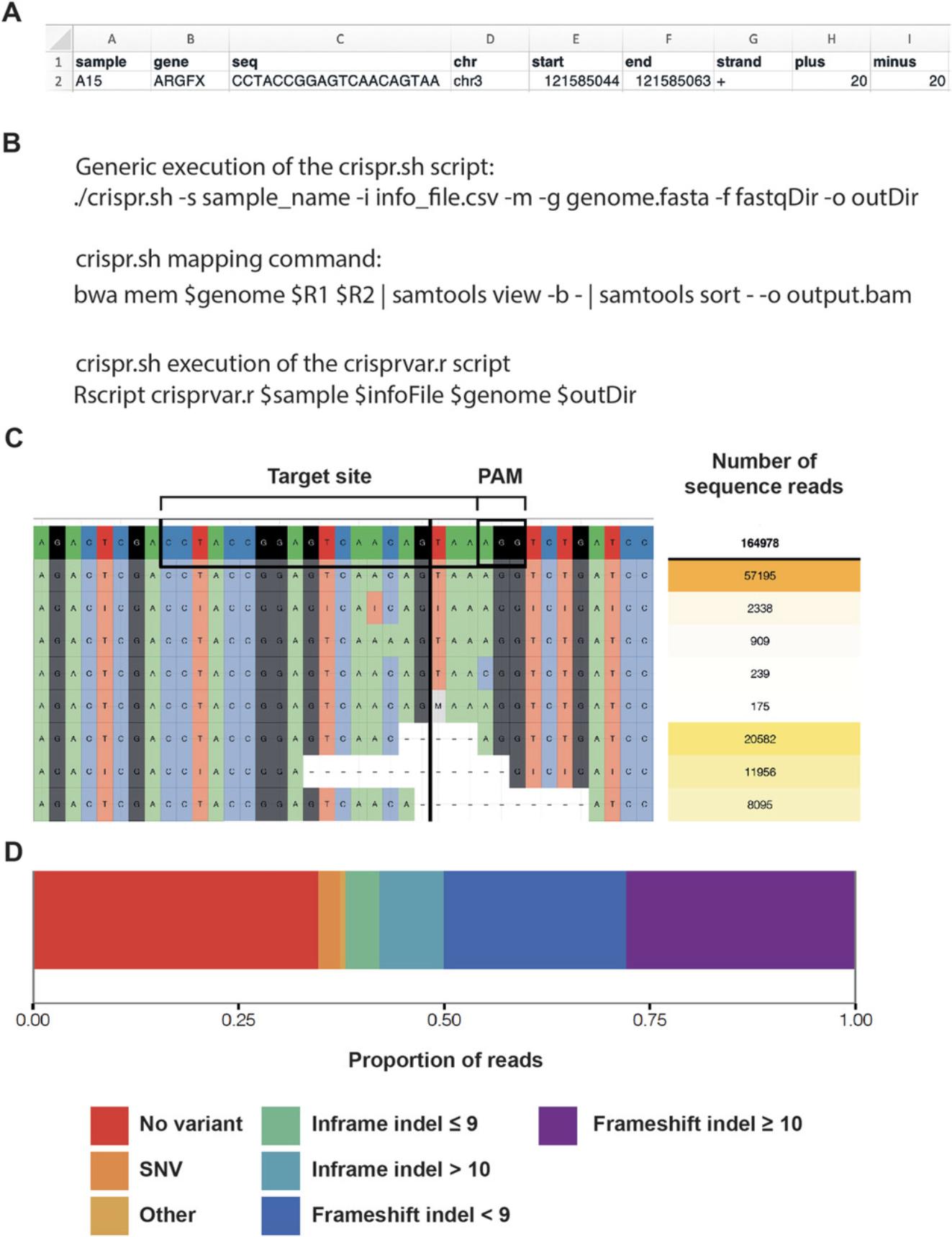
crispr.sh This script first optionally executes the mapping of MiSeq reads to the desired reference genome (Fig. 6B). Following mapping, the script directs to the following crisprvar.r script to run the analysis pipeline itself. To execute this script, one must provide the following parameters:
- 1.The sample name and info_file.csv.
- 2.Whether to proceed with mapping the MiSeq reads (to perform mapping, include the relevant flag).
- 3.A file path to the desired reference genome. This directory must include both an FAI index and a BWA index of the genome.
- 4.The input directory, while will contain the appropriate trimmed and corrected .fastq files that were generated by correct_fastq.sh.
- 5.The output directory for the BAM files, plots, and tables.
These BAM files can be viewed in a typical genome browser (e.g., IGV) and manually assessed for correct mapping. Additionally, the coverage of reads detected around the CRISPR-Cas9 target site will sometimes indicate the prevalence of indel mutations, for instance, a large number of mutations centered on the expected cut site would lead to reduced coverage at that position.
crisprvar.r This R script performs the actual analysis of indel presence within the trimmed and corrected MiSeq reads. The script reads all of the necessary variable parameters from within the crispr.sh script and so does not require modification between sample runs. Ultimately, this analysis provides a number of outputs (Fig. 6C and 6D), including:
- Plots (as PDFs):
- ○ "sample_name".pdf – the top 50 variants in the region around the CRISPR-Cas9 target site;
- ○ "sample_name"_pie.pdf – pie charts displaying the type and proportion of the most common indels; and,
- ○ "sample_name"_allele_freqs.pdf – box plots displaying the proportion of different types of variants, e.g., frameshift <10 bp, in-frame <10 bp.
- Tables (as text files):
- ○ "sample_name"_mutEffic.txt – the overall mutation efficiency within a sample, including basic statistics and read counts for wild-type and edited categories; and,
- ○ "sample_name"_variants.txt – a list of all of the variants occurring in the region around the CRISPR-Cas9 target site.
Basic Protocol 6–Karyotype Analysis
Align_lowpassWGS.sh This script enables a straightforward alignment of reads generated by low-pass WGS to the human hg19 genome (or your preferred build) via BWA. To execute the script, one needs to:
- 1.Define the file path to the directory containing the low-pass WGS .fastq files in line #8;
- 2.Define the desired reference genome for mapping in line #12; and,
- 3.Define the desired file path for the output of BAM files in line #15.
Proper execution of this script will yield the BAM files needed as input for the next step.
Generate_CNP.R This script enables the generation of visual copy number profiles (CNPs) from low-pass WGS data from single samples, within an R environment. To execute the script, one needs to:
- 1.Define the name of the first sample to be analyzed in line #8; and,
- 2.Define the file path relating to the output of the previous script, i.e., the directory containing the sorted BAM files from mapping, in line #10.
Other parameters can be adjusted depending on the specific requirements of the user, including:
- The size (x) of the genomic “bins” used for the analysis in line #12, getBinAnnotations(binSize = x) – example is 100 kb.
- The opportunity to generate separate plots for individual chromosomes from line #28 onwards – the example shows how to generate separate plots for chromosomes 5, 6, and 7.
Understanding Results
Basic Protocol 1 outlines the process of designing sgRNAs and cloning them into a suitable bacterial plasmid
During transformation, a number of colonies should form, as they should be ampicillin resistant due to uptake of the plasmid. Sequencing of these colonies should then return the 5′ - CACCG – Nx20 – GTTT – 3′ sequence. From this, a glycerol stock of the transformed bacteria can then be generated and stored indefinitely at −80°C.
Good sgRNAs should be unique gene regions with a minimal likelihood of binding to off-target sequences. sgRNAs with off-target sites harboring zero base pair mismatches (i.e., the sgRNA is completely complementary to an off-target locus) should always be discarded. sgRNAs with off-target sites harboring one mismatch (i.e., the sgRNA and off-target have one base pair dissimilarity) can be tolerable if the mismatch is within 10 bp of the PAM sequence (the ‘seed’ sequence). Off-target sites with >3 bp mismatches to the sgRNA are generally of no concern.
Midipreps from a 50 ml suspension of bacteria should generate approximately 100 μg of DNA. This plasmid DNA should then be concentrated to ∼4000 ng/μl for transfection into hPSCs. Concentrations lower than this will not be efficiently transfected into hPSCs.
Basic Protocol 2 is the earliest predictor of guide efficiency
The T7 DNA gel should result in a clear and distinct band at roughly 100 bp length. The T7 DNA should be around 100-300 ng/μl in concentration with a 260/280 nm ratio of >1.8. Measurements that are below this, or an issue with the T7 100 bp band, would suggest the T7 PCR is inefficient. To improve this step, perform a temperature gradient of the T7 primers.
The IVT RNA product should result in a relatively clear and distinct band at roughly 100 bp in length. The RNA product should be around 100-300 ng/μl in concentration with a 260/280 nm ratio of >1.8 and a 260/230 nm ratio of >2.0. The lack of a band would suggest an issue in the T7 PCR or the handling of the RNA, while a highly smeared band would suggest issues with an RNase-free environment that need to be resolved as above.
The IVC using an efficient sgRNA should result in the formation of two distinct bands of lengths equal to the predicted cleavage products. The presence of three weak bands with one of the same size as the uncleaved product would be indicative of a poor sgRNA on target cleavage. Increasing the Cas9 protein concentration could improve rates of cleavage or other sgRNAs could be used.
Basic Protocol 3 outlines the process of nucleofecting cells
After initial plating, it will be difficult to identify hPSCs among the MEFs. In the 2 days of puromycin selection, there should be a moderate-to-high amount of cell death, particularly in the GFP control well as these lack puromycin resistance. Over the next 2-4 days after selection, visible colonies of compact hPSCs should begin to emerge in a handful of locations across the plate.
Basic Protocol 4 will generate a set of MiSeq data detailing the presence and nature of indel mutations
The MiSeq data will be presented as the sequences observed in next-generation sequencing data along with the frequency with which the sequence is observed. The analysis pipeline will collapse reads according to sequence similarity and enable the calculation of frequency of out of frame mutations.
Basic Protocol 5 will produce 12 clonal targeted hPSC populations and Basic Protocol 6 will generate a CNP to identify chromosomal abnormalities
The CNP data will be presented as the number of times a particular sequence was observed. In this way, a wild-type 46 chromosome XX sample would have 2 copies for each sequence. However, a ratio greater than or less than two at certain loci or entire chromosomes suggests the presence of chromosome aneuploidies, with gain or loss of DNA, respectively.
Time Considerations
The in silico prediction of guides can be done in 1-2 days, but perhaps more if a thorough review of literature and similar sequences is needed.
Ligating oligonucleotides into bacteria takes 3 days, with most of the time needed to grow bacteria.
In vitro transcription reactions take around 5-6 hr. However, refining the technique and getting RNA to an acceptable quality for those inexperienced, along with working with few samples at a time to minimize RNA contamination, means it can take ∼1 week to generate an acceptable sgRNA sample.
Amplicon generation takes approximately 1 week to design, test and sequence amplicons generated. If there are issues and primers must be redesigned, it could take up to 2 weeks.
In vitro cleavage experiments take 2-3 hr to perform.
Nucleofection with a Lonza nucleofector takes approximately 1-2 hr depending on number of samples. The following selection and expansion of cells is between 1 and 2 weeks depending on when the time points of collection are.
Preparing samples for MiSeq analysis takes about 3 hr, including the time to extract DNA and run the PCR reaction.
Outsourced MiSeq analysis should take 1-3 weeks to return data. Subsequent analysis using the provided pipeline takes less than an hour.
Selecting colonies for single-cell cloning should take 1-2 hr. Subsequent growing of colonies to workable populations from single cells takes approximately 1-2 weeks.
Acknowledgements
The authors thank all members of the Niakan lab for their technical assistance and for help and comments on the paper. The authors are grateful to the Francis Crick Institute's Science Technology Platforms: Robert Goldstone and Deb Jackson from the Advanced Sequencing Facility and Lyn Healy, Liani Gasparni Devito and Elspeth Gamble from the Human Embryo and Stem Cell Unit. The authors thank Benjamin Fletcher for providing Figure 4D. Work in the Niakan and Turner laboratories is supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001120, FC001193), the UK Medical Research Council (FC001120, FC001193), and the Wellcome Trust (FC001120, FC001193). Work in the Turner laboratory is funded by the European Research Council (CoG 647971). For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Author Contributions
Oliver J. Bower : investigation, methodology, validation, writing original draft, writing review and editing; Afshan McCarthy : conceptualization, investigation, methodology, validation, writing-review and editing; Rebecca A. Lea : investigation, methodology, writing review and editing; Gregorio Alanis-Lobato : formal analysis, methodology; Jasmin Zohren : formal analysis, methodology, writing review and editing; Claudia Gerri : formal analysis, methodology, writing review and editing; James M.A. Turner : funding acquisition, project administration, resources, supervision, writing review and editing; Kathy K. Niakan : conceptualization, funding acquisition, project administration, resources, supervision, writing review and editing
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Literature Cited
- 1000 Genomes Project Consortium. (2015) A global reference for human genetic variation. Nature , 526, 68–74. doi: 10.1038/nature15393.
- Adikusuma, F., Piltz, S., Corbett, M. A., Turvey, M., McColl, S. R., Helbig, K. J., … Thomas, P. Q. (2018). Large deletions induced by Cas9 cleavage. Nature , 560(7717), E8–E9. doi: 10.1038/s41586-018-0380-z.
- Ahuja, A. K., Jodkowska, K., Teloni, F., Bizard, A. H., Zellweger, R., Herrador, R., … Lopes, M. (2016). A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nature Communications , 7(1), 10660. doi: 10.1038/ncomms10660.
- Avior, Y., Sagi, I., & Benvenisty, N. (2016). Pluripotent stem cells in disease modelling and drug discovery. Nature Reviews Molecular Cell Biology , 17(3), 170–182. doi: 10.1038/nrm.2015.27.
- Biasio, A. D., Vrana, J. A., Zhou, P., Qian, L., Bieszczad, C. K., Braley, K. E., … Craig, R. W. (2007). N-terminal truncation of antiapoptotic MCL1, but not G2/M-induced phosphorylation, is associated with stabilization and abundant expression in tumor cells. Journal of Biological Chemistry , 282(33), 23919–23936. doi: 10.1074/jbc.M700938200.
- Bibikova, M., Carroll, D., Segal, D. J., Trautman, J. K., Smith, J., Kim, Y. G., & Chandrasegaran, S. (2001). Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Molecular and Cellular Biology , 21(1), 289–297. doi: 10.1128/MCB.21.1.289-297.2001.
- Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., … Bonas, U. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science , 326(5959), 1509–1512. doi: 10.1126/science.1178811.
- Brinkman, E. K., Chen, T., Amendola, M., & van Steensel, B. (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Research , 42(22), e168–e168. doi: 10.1093/nar/gku936.
- Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., & Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods , 13(7), 581–583. doi: 10.1038/nmeth.3869.
- Capecchi, M. R. (1989). Altering the genome by homologous recombination. Science , 244(4910), 1288–1292. doi: 10.1126/science.2660260.
- Cohen, D. E., & Melton, D. (2011). Turning straw into gold: Directing cell fate for regenerative medicine. Nature Reviews Genetics , 12(4), 243–252. doi: 10.1038/nrg2938.
- Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., … Zhang, F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science , 339(6121), 819–823. doi: 10.1126/science.1231143.
- Cullot, G., Boutin, J., Toutain, J., Prat, F., Pennamen, P., Rooryck, C., … Bedel, A. (2019). CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nature Communications , 10(1), 1136. doi: 10.1038/s41467-019-09006-2.
- Dehairs, J., Talebi, A., Cherifi, Y., & Swinnen, J. V. (2016). CRISP-ID: Decoding CRISPR mediated indels by Sanger sequencing. Scientific Reports , 6, 28973. doi: 10.1038/srep28973.
- Doench, J. G., Fusi, N., Sullender, M., Hegde, M., Vaimberg, E. W., Donovan, K. F., … Root, D. E. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature Biotechnology , 34(2), 184–191. doi: 10.1038/nbt.3437.
- Draper, J. S., Smith, K., Gokhale, P., Moore, H. D., Maltby, E., Johnson, J., … Andrews, P. W. (2004). Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nature Biotechnology , 22(1), 53–54. doi: 10.1038/nbt922.
- Eguizabal, C., Aran, B., Chuva de Sousa Lopes, S. M., Geens, M., Heindryckx, B., Panula, S., … Veiga, A. (2019). Two decades of embryonic stem cells: A historical overview. Human Reproduction Open , 2019, hoy024. doi: 10.1093/hropen/hoy024.
- Elbing, K. L., & Brent, R. (2018). Recipes and tools for culture of Escherichia coli. Current Protocols in Molecular Biology , 125, e83.doi: 10.1002/cpmb.83.
- Elbing, K. L., & Brent, R. (2019). Growth of E. coli on solid media. Current Protocols in Molecular Biology , 125, e82. doi: 10.1002/cpmb.82.
- Elbing, K. L., & Brent, R. (2019). Growth of E. coli on solid media. Current Protocols in Molecular Biology , 125, e82. doi:10.1002/cpmb.82.
- Elitt, M. S., Barbar, L., & Tesar, P. J. (2018). Drug screening for human genetic diseases using iPSC models. Human Molecular Genetics , 27(R2), R89–R98. doi: 10.1093/hmg/ddy186.
- Elliott, A. M., Elliott, K. A. H., & Kammesheidt, A. (2010). High resolution array-CGH characterization of human stem cells using a stem cell focused microarray. Molecular Biotechnology , 46(3), 234–242. doi: 10.1007/s12033-010-9294-1.
- Engler, C., Gruetzner, R., Kandzia, R., & Marillonnet, S. (2009). Golden gate shuffling: A one-pot DNA shuffling method based on type IIs restriction enzymes. PloS One , 4(5)doi: 10.1371/journal.pone.0005553.
- Firth, A. L., Menon, T., Parker, G. S., Qualls, S. J., Lewis, B. M., Ke, E., … Verma, I. M. (2015). Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Reports , 12(9), 1385–1390. doi: 10.1016/j.celrep.2015.07.062.
- Fogarty, N. M. E., McCarthy, A., Snijders, K. E., Powell, B. E., Kubikova, N., Blakeley, P., … Niakan, K. K. (2017). Genome editing reveals a role for OCT4 in human embryogenesis. Nature , 550(7674), 67–73. doi: 10.1038/nature24033.
- Garitaonandia, I., Amir, H., Boscolo, F. S., Wambua, G. K., Schultheisz, H. L., Sabatini, K., … Laurent, L. C. (2015). Increased risk of genetic and epigenetic instability in human embryonic stem cells associated with specific culture conditions. PloS One , 10(2), e0118307. doi: 10.1371/journal.pone.0118307.
- Guo, G., Stirparo, G. G., Strawbridge, S. E., Spindlow, D., Yang, J., Clarke, J., … Smith, A. (2021). Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell , 28(6), 1040–1056.e6. doi: 10.1016/j.stem.2021.02.025.
- Hsu, P. D., Lander, E. S., & Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell , 157(6), 1262–1278. doi: 10.1016/j.cell.2014.05.010.
- Huang, X., Wang, Y., Yan, W., Smith, C., Ye, Z., Wang, J., … Cheng, L. (2015). Production of gene-corrected adult beta globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem Cells , 33(5), 1470–1479. doi: 10.1002/stem.1969.
- Ilie, L., & Molnar, M. (2013). RACER: Rapid and accurate correction of errors in reads. Bioinformatics , 29(19), 2490–2493. doi: 10.1093/bioinformatics/btt407.
- International Stem Cell Initiative, Amps, K., Andrews, P. W., Anyfantis, G., Armstrong, L., Avery, S., … Zhou, Q. (2011). Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nature Biotechnology , 29(12), 1132–1144. doi: 10.1038/nbt.2051.
- Jacków, J., Guo, Z., Hansen, C., Abaci, H. E., Doucet, Y. S., Shin, J. U., … Christiano, A. M. (2019). CRISPR/Cas9-based targeted genome editing for correction of recessive dystrophic epidermolysis bullosa using iPS cells. Proceedings of the National Academy of Sciences , 116(52), 26846–26852; doi: 10.1073/pnas.1907081116.
- Jiang, F., & Doudna, J. A. (2017). CRISPR-Cas9 structures and mechanisms. Annual Review of Biophysics , 46, 505–529. doi: 10.1146/annurev-biophys-062215-010822 Epub 2017 Mar 30.
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., & Charpentier, E. (2012). A programmable dual RNA-guided DNA endonuclease in adaptive bacterial immunity. Science , 337(6096), 816–821. doi: 10.1126/science.1225829.
- Jinek, M., East, A., Cheng, A., Lin, S., Ma, E., & Doudna, J. (2013). RNA-programmed genome editing in human cells. Elife , 2, e00471. doi: 10.7554/eLife.00471.
- Kim, Y. G., Cha, J., & Chandrasegaran, S. (1996). Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proceedings of the National Academy of Sciences of the United States of America , 93(3), 1156–1160.
- Kim, D., Bae, S., Park, J., Kim, E., Kim, S., Yu, H. R., … Kim, J.-S. (2015). Digenome-seq: Genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nature Methods , 12(3), 237–243. 1 p following 243. doi: 10.1038/nmeth.3284.
- Kim, D., Kim, S., Kim, S., Park, J., & Kim, J.-S. (2016). Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Research , 26(3), 406–415. doi: 10.1101/gr.199588.115.
- Kosicki, M., Tomberg, K., & Bradley, A. (2018). Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nature Biotechnology , 36(8), 765–771. doi: 10.1038/nbt.4192.
- Lee, K., Cho, H., Rickert, R. W., Li, Q. V., Pulecio, J., Leslie, C. S., & Huangfu, D. (2019). FOXA2 is required for enhancer priming during pancreatic differentiation. Cell Reports , 28(2), 382–393.e7. doi: 10.1016/j.celrep.2019.06.034.
- Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics (Oxford, England) , 27(21), 2987–2993. doi: 10.1093/bioinformatics/btr509.
- Li, H., & Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics , 26(5), 589–595. doi: 10.1093/bioinformatics/btp698.
- Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., … Durbin, R. (2009). The sequence alignment/map format and SAMtools. Bioinformatics , 25(16), 2078–2079. doi: 10.1093/bioinformatics/btp352.
- Lindsay, H., Burger, A., Biyong, B., Felker, A., Hess, C., Zaugg, J., … Robinson, M. D. (2016). CrispRVariants charts the mutation spectrum of genome engineering experiments. Nature Biotechnology , 34(7), 701–702. doi: 10.1038/nbt.3628.
- Liu, X., Tan, J. P., Schröder, J., Aberkane, A., Ouyang, J. F., Mohenska, M., … Polo, J. M. (2021). Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature , 591(7851), 627–632. doi: 10.1038/s41586-021-03372-y.
- Ludwig, T., & A Thomson, J. (2007). Defined, feeder-independent medium for human embryonic stem cell culture. Current Protocols in Stem Cell Biology , 2, 1C.2.1−1C.2.16. doi: 10.1002/9780470151808.sc01c02s2.
- Ma, Y., Brewer, J. W., Alan Diehl, J., & Hendershot, L. M. (2002). Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. Journal of Molecular Biology , 318(5), 1351–1365. doi: 10.1016/S0022-2836(02)00234-6.
- Malhotra, J. D., & Kaufman, R. J. (2007). The endoplasmic reticulum and the unfolded protein response. Seminars in Cell & Developmental Biology, 18(6), 716–731. doi: 10.1016/j.semcdb.2007.09.003.
- Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., … Church, G. M. (2013). RNA-guided human genome engineering via Cas9. Science , 339(6121), 823–826. doi: 10.1126/science.1232033.
- Martello, G., & Smith, A. (2014). The nature of embryonic stem cells. Annual Review of Cell and Developmental Biology , 30(1), 647–675. doi: 10.1146/annurev-cellbio-100913-013116.
- McIntire, E., Leonhard, K., Taapken, S., & Larson, A. L. (2021). G-banded karyotyping of human pluripotent stem cell cultures. Methods in Molecular Biology (Clifton, N.J.) , 2239, 251–268. doi: 10.1007/978-1-0716-1084-8_16.
- Moris, N., Anlas, K., van den Brink, S. C., Alemany, A., Schröder, J., Ghimire, S., … Martinez Arias, A. (2020). An in vitro model of early anteroposterior organization during human development. Nature , 582(7812), 410–415. doi: 10.1038/s41586-020-2383-9.
- Murry, C. E., & Keller, G. (2008). Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell , 132(4), 661–680. doi: 10.1016/j.cell.2008.02.008.
- Ng, H.-H., & Surani, M. A. (2011). The transcriptional and signalling networks of pluripotency. Nature Cell Biology , 13(5), 490–496. doi: 10.1038/ncb0511-490.
- Nichols, J., & Smith, A. (2009). Naive and primed pluripotent states. Cell Stem Cell , 4(6), 487–492. doi: 10.1016/j.stem.2009.05.015.
- Niwa, H. (2007). How is pluripotency determined and maintained? Development , 134(4), 635–646. doi: 10.1242/dev.02787.
- Owens, D. D. G., Caulder, A., Frontera, V., Harman, J. R., Allan, A. J., Bucakci, A., … de Bruijn, M. F. T. R. (2019). Microhomologies are prevalent at Cas9-induced larger deletions. Nucleic Acids Research , 47(14), 7402–7417. doi: 10.1093/nar/gkz459.
- Phelan, K., & May, K. M. (2015). Basic techniques in mammalian cell culture. Current Protocols in Cell Biology , 66, 1.1.1–1.1.22.doi: 10.1002/0471143030.cb0101s66.
- Przewrocka, J., Rowan, A., Rosenthal, R., Kanu, N., & Swanton, C. (2020). Unintended on-target chromosomal instability following CRISPR/Cas9 single gene targeting. Annals of Oncology , 31(9), 1270–1273. doi: 10.1016/j.annonc.2020.04.480.
- Rivron, N. C., Frias-Aldeguer, J., Vrij, E. J., Boisset, J.-C., Korving, J., Vivié, J., … Geijsen, N. (2018). Blastocyst-like structures generated solely from stem cells. Nature , 557(7703), 106–111. doi: 10.1038/s41586-018-0051-0.
- Rossi, A., Kontarakis, Z., Gerri, C., Nolte, H., Hölper, S., Krüger, M., & Stainier, D. Y. (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature , 524(7564), 230–233. doi: 10.1038/nature14580.
- Seol, J.-H., Shim, E. Y., & Lee, S. E. (2018). Microhomology-mediated end joining: Good, bad and ugly. Mutation Research , 809, 81–87. doi: 10.1016/j.mrfmmm.2017.07.002.
- Sfeir, A., & Symington, L. S. (2015). Microhomology-mediated end joining: A back-up survival mechanism or dedicated pathway? Trends in Biochemical Sciences , 40(11), 701–714. doi: 10.1016/j.tibs.2015.08.006.
- Shahbazi, M. N., Scialdone, A., Skorupska, N., Weberling, A., Recher, G., Zhu, M., … Zernicka-Goetz, M. (2017). Pluripotent state transitions coordinate morphogenesis in mouse and human embryos. Nature , 552(7684), 239–243. doi: 10.1038/nature24675.
- Shin, H. Y., Wang, C., Lee, H. K., Yoo, K. H., Zeng, X., Kuhns, T., … Hennighausen, L. (2017). CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nature Communications , 8, 15464. doi: 10.1038/ncomms15464.
- Stephenson, E., Ogilvie, C. M., Patel, H., Cornwell, G., Jacquet, L., Kadeva, N., … Ilic, D. (2010). Safety paradigm: Genetic evaluation of therapeutic grade human embryonic stem cells. Journal of the Royal Society, Interface , 7(Suppl 6), S677–688. doi: 10.1098/rsif.2010.0343.focus.
- Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., & Yamanaka, S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell , 131(5), 861–872. doi: 10.1016/j.cell.2007.11.019.
- Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., & Jones, J. M. (1998). Embryonic stem cell lines derived from human blastocysts. Science , 282(5391), 1145–1147. doi: 10.1126/science.282.5391.1145.
- Tsai, S. Q., Zheng, Z., Nguyen, N. T., Liebers, M., Topkar, V. V., Thapar, V., … Joung, J. K. (2015). GUIDE-Seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature Biotechnology , 33(2), 187–197. doi: 10.1038/nbt.3117.
- Tsai, S. Q., Nguyen, N. T., Malagon-Lopez, J., Topkar, V. V., Aryee, M. J., & Joung, J. K. (2017). CIRCLE-seq: A highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nature Methods , 14(6), 607–614. doi: 10.1038/nmeth.4278.
- Voytas, D. (2001). Agarose gel electrophoresis. Current Protocols in Molecular Biology , 51, 2.5A.1−2.5A.9.
- Wang, Q., Zou, Y., Nowotschin, S., Kim, S. Y., Li, Q. V., Soh, C.-L., … Massagué, J. (2017). The p53 family coordinates Wnt and nodal inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell , 20(1), 70–86. doi: 10.1016/j.stem.2016.10.002.
- Warmflash, A., Sorre, B., Etoc, F., Siggia, E. D., & Brivanlou, A. H. (2014). A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nature Methods , 11(8), 847–854. doi: 10.1038/nmeth.3016.
- Weissbein, U., Peretz, M., Plotnik, O., Yanuka, O., Sagi, I., Golan-Lev, T., & Benvenisty, N. (2019). Genome-wide screen for culture adaptation and tumorigenicity-related genes in human pluripotent stem cells. IScience , 11, 398–408. doi: 10.1016/j.isci.2018.12.031.
- Wu, H., Kim, K. J., Mehta, K., Paxia, S., Sundstrom, A., Anantharaman, T., … Teitell, M. A. (2008). Copy number variant analysis of human embryonic stem cells. Stem Cells , 26(6), 1484–1489. doi: 10.1634/stemcells.2007-0993.
- Wu, X., Kriz, A. J., & Sharp, P. A. (2014). Target specificity of the CRISPR-Cas9 system. Quantitative Biology , 2(2), 59–70. doi: 10.1007/s40484-014-0030-x.
- Yanagida, A., Spindlow, D., Nichols, J., Dattani, A., Smith, A., & Guo, G. (2021). Naive stem cell blastocyst model captures human embryo lineage segregation. Cell Stem Cell , 0(0), 1016–1022. doi: 10.1016/j.stem.2021.04.031.
- Yu, L., Wei, Y., Duan, J., Schmitz, D. A., Sakurai, M., Wang, L., … Wu, J. (2021). Blastocyst-like structures generated from human pluripotent stem cells. Nature , 591(7851), 620–626. doi: 10.1038/s41586-021-03356-y.
- Zeman, M. K., & Cimprich, K. A. (2014). Causes and consequences of replication stress. Nature Cell Biology , 16(1), 2–9. doi: 10.1038/ncb2897.
- Zuccaro, M. V., Xu, J., Mitchell, C., Marin, D., Zimmerman, R., Rana, B., … Egli, D. (2020). Allele-specific chromosome removal after Cas9 cleavage in human embryos. Cell , 183(6), 1650–1664.e15. doi: 10.1016/j.cell.2020.10.025.
Citing Literature
Number of times cited according to CrossRef: 2
- Elie Frank, Michel Cailleret, Constantin Nelep, Pascal Fragner, Jérome Polentes, Elise Herardot, Lina El Kassar, Karine Giraud-Triboult, Christelle Monville, Karim Ben M’Barek, Semi-automated optimized method to isolate CRISPR/Cas9 edited human pluripotent stem cell clones, Stem Cell Research & Therapy, 10.1186/s13287-023-03327-2, 14 , 1, (2023).
- Tatiana V. Nikitina, Igor N. Lebedev, Stem Cell-Based Trophoblast Models to Unravel the Genetic Causes of Human Miscarriages, Cells, 10.3390/cells11121923, 11 , 12, (1923), (2022).

