FPCount protocol - Short protocol
Eszter Csibra, Guy-Bart Stan
Abstract
FPCount is a complete protocol for fluorescent protein calibration, consisting of:
-
FP expression/purification using Thermo's HisPur Cobalt Resin.
-
FP concentration determination in a microplate reader.
-
FP fluorescence quantification in a microplate reader.
Results can be analysed with the corresponding R package, FPCountR.
This short version uses the ECmax protein quantification protocol, and is only suitable for FPs with entries in FPbase. If you want to verify or validate results, it's recommended you follow the complete protocol, which describes three protein quantification methods. The short protocol also skips the SDS-PAGE steps. If you require these, please see the complete protocol.
Summary
-
Expression
-
Harvesting/Washing
-
Lysis
-
Fractionation
-
Purification
-
Protein concentration and buffer exchange
-
Quantification of FP concentration (part1)
-
Quantification of FP fluorescence
-
Protein storage
-
Calibration of Plate Reader
Before start
This short version of the protocol uses the ECmax protein quantification protocol, and is only suitable for FPs with entries in FPbase. If you want to verify or validate results, it's recommended you follow the complete protocol, which describes three protein quantification methods. The short protocol also skips over the DNase step, and the SDS-PAGE steps. If you require these steps, please see the complete protocol.
Steps
Expression
[ Day 1 ]
Overnight culture set-up:
- 50ml LB
- 50ul cam
- 50ul arabinose 0.02%
- glycerol stock scraping of BL21/pS381_ara_His-FP transformant (or equivalent)
- 30oC 250rpm
- overnight expression…
Harvesting and Washing
[ Day 2 ]
Buffers:
- Wash buffer = T50N150
- 50 mM Tris-Cl pH 7.5, 150 mM NaCl
- Doesn’t need protease inhibitors
- Can be substituted by T50N300
- Resuspension buffer = T50N300+pi
- 50 mM Tris-Cl pH 7.5, 300 mM NaCl
- protease inhibitors (pi; 1 tablet/10ml)
- filter sterilise as pi makes things cloudy/doesn’t go into solution well
- Lysis Buffer = T50N300+pi
- lysozyme 1X (100ug/ml)
- DNase I (1000U/ml)
- CaCl2 stock
- MgCl2 stock
- 2X Laemmli’s buffer (2xLB)
- Binding Buffer (BB) = T50N300+pi+10imid
- T50N300+pi, 10 mM imidazole
- Elution Buffer (EB) = T50N300+pi+150imid
- T50N300+pi, 150 mM imidazole
- Dilution Buffer = T5N15+pi
- For protein assays
- How much buffer will I need?
- T50N150 - maybe 35ml/FP
- T50N300 + pi (f/s) - make master stock > 10ml per FP.
- T50N300 + pi + lysozyme = 5ml per FP
- T50N300 + pi + 10mM imid = 4ml per FP
- T50N300 + pi + 150mM imid = 1ml per FP
- T5N15 + pi = {5ml for BSA + 8ml for each FP}
- microBCA working reagent (WR) = 12ml per plate (plate = 3 FPs) OR {3ml for BSA + 3ml per FP}
- total pi tablets required = (18ml per FP + 5ml BSA) = </= 23ml per FP = </= 3 pi per FP
Procedure:
- Prechill 1x 50ml falcon tube per FP on ice, 15min
- Prechill 1-2x 50ml falcon tubes per FP on ice - for sonication (choose 1x aliquot if using 20 OD cells, or 2x for 40 OD cells)
- Prechill big centrifuge, 4oC
- Remove culture from incubator; for some FPs it will be clear by eye if expression levels are good.
- Transfer to falcon on ice; cool for 20min
- From now on cultures and protein should be kept on ice and spun at 4oC unless otherwise stated.
- Take OD (use 100ul of culture 1:10 in LB)
- Expect maybe 4-6 OD/ml.
- Calculate volume or fraction of total required for 20 or 40 OD worth of cells. (20 OD = 1x 20OD/2ml aliquot for sonication; 40 OD = 2x 20OD/2ml aliquot for sonication)
- Expect fraction to be 0.1-0.2 meaning we’re only using 10-20% of the culture for this purification even if we get through the whole 40 OD (which would require loading 7* 600ul on the spin columns). So expecting vol to be 5-10ml.
Example OD calculation:
| A | B | C | D | E | F | G |
|---|---|---|---|---|---|---|
| 40/total OD | 40/total * 50 | |||||
| 1 | mCherry | 0.418 | 4.18 | 209 | 0.19 | 9.57 |
| 2 | eg1 | 0.3 | 3 | 150 | 0.267 | 13.3 |
| 3 | eg2 | 0.6 | 6 | 300 | 0.133 | 6.67 |
- Add 20 or 40 OD to the prechilled tubes set aside for aliquotted cultures.
- (Original cultures can stay on ice or be stored in fridge.)
- Spin 3,220xg, 10min, 4oC
- Resuspend in 5ml WASH buffer w pipetboy/5ml stripette; Add 30ml more WASH buffer or so
- Spin 3,220xg, 10min, 4oC
- Resuspend in 2ml (for 20 OD) or 4ml (for 40 OD) Lysis Buffer.
- Lysis Buffer = T50N300 + pi + lysozyme. (No DTT for His tag purifs.)
- eg. for 5 purifications, Take 22 ml of T50N300+pi and add 44ul lysozyme.
- If using 40 OD, split cells into 2x falcons of 20OD/2ml each.
Lysis
Prep for next stage: pre-chill the microfuge to 4oC.
Lysis by Sonication
- Stand falcon in small plastic beaker full of ice.
- Sonicate: 50% amplitude, 10s on/off, 2min .
- NB. 2min means 2min of sonication. as we’re doing 10s on/off, this takes 4min.
- Solution should go from turbid to clear.
- If doing 40 OD, you will have two falcons.
- If you have many FPs, after 6 falcons you have enough sample to fill the microfuge for the next stage - it’s worth starting the DNase step (30min) then the spin (30min) before the other samples are sonicated.
DNase I treatment
Optional step: essential if using A280 assay but not essential for the ECmax assay. Note that DNase I = 31 kDa meaning similar in size to FPs in a way that would affect estimates in Gel1 (though not Gel2 as it shouldn’t bind the column), and is sensitive to vortexing.
- Prepare DNase I stock: 1000 U/ml DNase I in ddH2O
- To lysates in T50N300, add:
- DNase I to 50 U/ml final (20X dilution)
- CaCl2 to 5mM final (13mM ideal for DNase I, <5mM recommended with His resins)
- MgCl2 to 50mM final
- Mix thoroughly
- Reaction: 30min at 4oC
Fractionation
Prep for next stage: prep eppies and cool them for after the spin
[ Day 2 ]
- Spin out insoluble fraction.
- Split 2ml lysates into 4x 0.5ml prechilled eppies.
- Spin in prechilled microfuge - 30min 16Kg 4oC.
- Result: 4x 0.5ml of soluble lysate (if using 20 OD) or 8x 0.5ml soluble lysate (if using 40 OD)
- Transfer SOLUBLE fractions to new tubes - can mix so that you have fewer eppies.

Prep for next step: Change temp on microfuge to 21oC, and open lid to let it warm up. If incubations are done at RT to speed up kinetics, they should be spun at ~RT too. Proteins should be protected by the protease inhibitors.
Affinity Purification
There are a number of ways to purify His-tagged proteins. I opted for the HisPur Cobalt resin for a few reasons. Resins are available in agarose bead and magnetic bead format - magnetic beads are attractive for high throughput and automated protocols, but their binding capacities are 100-1000fold lower per volume than agarose resins. While I don’t need grams of protein, I did require ug-mg yields, so it had to be agarose. The most popular resin is the Nickel resin (NiNTA) but I opted for Cobalt because, while its capacity is lower this is not a problem at the scales I require, but its specificity is higher ensuring purer protein.
Back of the envelope calculation to illustrate:
- HisPur Cobalt resin.
- Capacity of the resin: 10mg/ml (lower than NiNTA but higher specificity).
- spin columns
- I use Pierce Spin Columns - Snap Cap. The standard/smallest one has a max volume of 300ul 50% resin, which means a total yield of 3mg protein can be obtained from Cobalt resins, or 20mg protein max from NiNTA. This is typically sufficient: assuming all protein ends up in one quant assay, we’re talking a starting conc for the highest conc in the plate would be (E1 neat: 3mg/100ul = 3000ug/100ul = 30ug/ul; E1/10 =) 3ug/ul. If I get a conc of 500ng/ul here I’m happy (6x less). Minimum is probably 100ng/ul (30x less).
Nickel and Cobalt resins tolerate most common reagents except EDTA (so there’s no EDTA in the lysis buffer). DTT is disputed - manual for HisPur says to avoid it completely. DTT use (1mM) was tested and found to make not much difference to purification yields. Meaning DTT 1mM shouldn’t negatively affect the columns. Subsequently however I didn’t use it because it interferes w microBCA assays, and FPs seem happy enough without (but many of them only have a single Cys anyway).
[ Day 2 contd ]
Resin
- Take the HisCobalt resin out of the fridge.
- Shake and gently vortex resin stock bottle to make sure it is well mixed.
- Take 600ul 50% resin -> add it to an assembled spin column (max. volume 600ul).
- Spin 1’ 1000g to remove storage buffer
- Discard flowthrough (fth).
Resin equilibration
-
Resin equilibration rounds:
-
cap
-
add 300ul Binding Buffer (BB; T50N300, pi, 10mM imidazole - no lysozyme)
-
flick to mix
-
uncap
-
spin 1’ 1000g
-
discard fth
-
Repeat 3X. I frequently incubate third equilibration 15’ RT.
| A | B |
|---|---|
| 1 | |
| 2 | |
| 3 |
Binding
– – – Remember to add 10mM imidazole to samples before adding to column! – – –
- Binding rounds:
- cap
- add 600ul FP lysate (in T50N300, pi, lysozyme, 10mM imidazole)
- flick to mix
- Bind at RT for 15min-90min
- flick to mix every 5-10min
- uncap
- 1’ 1000g
Keep Flowthrough from at least one Binding Round for gel2: transfer flowthrough (after spin) from standard elution tube to a 1.5ml eppy.
- Repeat 1X-7X.
Binding can be done with just one sample of 600ul, or the full 40 OD cells (4ml lysate ~ 6+ sets of 600ul). (Though column can saturate by the 5th round of loading.)
| A | B |
|---|---|
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 |
Washing
Prep for next stage: prep EB if not done. Put EB at RT if not done.
-
Washing rounds:
-
cap
-
add 300ul-600ul BB
-
flick to mix
-
uncap
-
1’ 1000g
-
discard fth
-
Repeat 5X.
Standard recommended washing is with 5 column volumes BB, as 5X with 1 col vol (300ul), but when doing a high number of binding rounds, I like to increase this to 10 col vols (eg. 5X 600ul). Can add 1M NaCl to increase stringency.
| A | B |
|---|---|
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 |
Elutions
-
Elution rounds:
-
cap
-
add 300ul Elution Buffer (EB; T50N300, pi, 150mM imidazole)
-
flick to mix
-
Elution incubation, 5’ RT
-
uncap and Transfer to Elution eppy (1.5ml)
-
1’ 1000g
-
Elutions to fridge.
-
Repeat 3X.
| A | B |
|---|---|
| 1 | |
| 2 | |
| 3 |
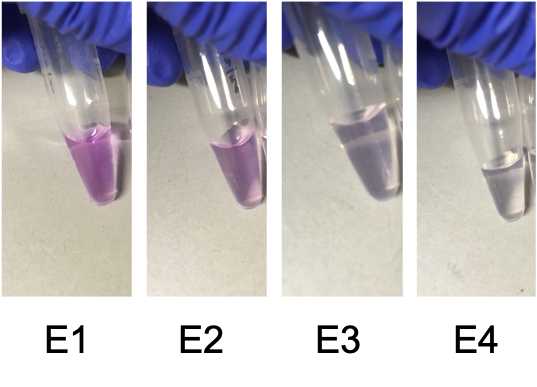
Protein concentration (not always optional)
[ Day 3 ]
I do a buffer exchange every time (to T5N15-+pi) because it is essential for microBCA and A280 assay accuracy. However, if only ECmax assays are going to be done, technically this step is optional. T5N15 + pi should be made fresh. There is no data on pi stability so in practise I never use >1day old buffer. Filter sterilise to get rid of cloudy precipitate (doesn’t disappear even when dissolved at 37oC. According to the manual, this is normal, but it may interfere with assays.)
Minimum concentrations for FP elutions in the next steps are > 100 ng/ul, as the highest conc to be measured in will be Elution/10, and 10 ng/ul is still within range of the protein assays. It usually makes sense to merge all elutions before starting.
Notes before starting:
- As E1>>E2>>E3, combining all three and concentrating back to the original volume of E1, 300ul, is unlikely to make a big difference in the conc of E1 pre-concentration to conc of E123 post-concentration. However, it should increase. But if this is further concentrated to 100ul then at least a 3X increase in concentration should result.
- Amicons claim 100 mM imidazole is the maximum compatibility of the columns. The elutions contain 150 mM. So technically the first step should be to DILUTE the elution 2-fold, but in practice it works fine as is.
- Amicons are not compatible with >0.1% Triton X100 and Triton can interfere w A280 absorbance in protein assays. If sonication was used, no problem. If Triton was used, typically this means there’s 0.1% in the lysis buffer. If the bound resin was washed with 10 column volumes of Triton-free buffer, perhaps Triton shouldn’t be an issue, but this hasn’t been tested.
- Amicon concentration can be taken as an opportunity for buffer exchange, eg. dilute out imidazole and protease inhibitors and buffer exchange from T50N300 to T5N15. But obviously to compare different proteins they should be treated the same. Note: for the experiments in the paper, I (i) merged and concentrated E1+E2+E3 (900ul) to 50ul, (ii) buffer exchanged (3x450ul) into T5N15 and made up to 100ul, (iii) split the FPs into 2x50ul and buffer exchanged (3x450ul) either into T5N15 again, or T5N15+pi. Step (ii) was required to remove imidazole, and step (iii) to add pi back to one half of the sample, because I wanted to compare T5N15 and T5N15+pi in the following assays. In practise, it makes sense to do only one buffer exchange, straight into your buffer of choice. Steps:
-
Situation: E1 is estimated at 100ng/ul
-
Plan:
-
- merge E1, E2, E3 (=300x3 = 900ul) and concentrate to 50ul
-
- buffer exchange and concentrate to < 100ul
-
- resuspend to 100-300ul
-
Use Amicon Ultra 10K columns
-
500ul capacity (500ul -> 15ul possible)
-
spin at 14Kg 10’ to concentrate
-
spin at 1Kg 1’ to recover
-
Step1. Concentration.
-
Add E1 (300ul) to 10K amicon column
-
5’ 14Kg spin at 21oC
-
expect it to go down to 100ul
-
discard flowthrough
-
Add E2 (300ul)
-
5’ 14Kg spin at 21oC
-
expect it to go down to 100ul
-
discard flowthrough
-
Add E3 (300ul)
-
10’ 14Kg spin at 21oC
-
expect it to go down to 50ul (needs to be <100ul)
-
Recover result (needs to be <50ul)
-
turn column over into fresh eppy
-
1’ 1000g
-
Take sample, measure volume precisely, dilute back to exactly 50ul (for single buffer exchange) or 100ul if needs to be split into two buffer exchanges (to compare buffers).
-
Step2. Buffer exchange into T5N15 -/+ protease inhibitors
-
Assuming we’re starting from 50ul protein:
-
Add E123 (50ul) to (fresh) 10K amicon column
-
10’ 14Kg spin at 21oC
-
expect it to go down to 50ul
-
discard flowthrough
-
Add 450ul buffer (1)
-
10’ 14Kg spin at 21oC
-
expect it to go down to 50ul
-
discard flowthrough
-
Add 450ul buffer (2)
-
10’ 14Kg spin at 21oC
-
expect it to go down to 50ul
-
discard flowthrough
-
Add 450ul buffer (3)
-
10’ 14Kg spin at 21oC
-
expect it to go down to 50ul
-
discard flowthrough
-
Recover result (needs to be <50ul)
-
turn column over into fresh eppy
-
1’ 1000g
-
Take sample, measure volume precisely, dilute back to 100ul+. 100ul is enough for one quantification, 200ul+ will allow for repeats.
FP Calibration in Plate Readers Protocol
The idea behind this protocol is to make most efficient use of protein. Therefore, ideally, one 100ul aliquot of the merged/concentrated Elution (E123), is all that is needed for all the fluorescence and concentration assays, and these can be done consecutively on a single dilution series. Currently an ‘exhaustive’ workflow includes 1 fluorescence assay and 3 protein assays. As this is the 'short' protocol, this only uses 1 fluorescence assay and 1 protein assay - the ECmax assay.
Summary:
- You will need 100ul elution of each FP to be calibrated.
- Access to all plate readers to be calibrated for a clear few hours is ideal, but the time required depends entirely on ambition: the number of instruments, channels and FPs.
- Prep dilutions as 200ul in the same type of plate as you use for bacterial assays -> absorbance scans and fluorescence quants.
Quantification of FP Concentration (part1)
There is only one protein concentration assay ('protein assay') in this simplified workflow - the ECmax assay.
Therefore plates to scan in order are:
- 200ul clear - ECmax assay/fluorescence assay
–
[ Day 3 contd ]
Prepare dilutions of FPs. Run scans in plate reader for FP quantification.
1. Make Protein Dilutions in Eppies
In order to validate the protein quantitation assays, I dilute each FP using serial dilutions and measure the concentration and fluorescence of each. In order to maximise both the dilution series and the number of FPs quantifiable per plate, I typically arrange FPs in 96-well plates in pairs of rows (AB, CD, EF, GH) - each FP is therefore measured as a duplicate. This leaves the columns for the dilution series itself, with 11 used for protein dilutions, and one for the buffer. (NB: In theory, arranging FPs in columns, with 7 dilutions plus one buffer for each FP, would allow for 6 proteins to be measured at the same time.)
I recommend preparing dilution series in 1.5ml eppies: they are easier to handle (and see into) than wells of a deep well plate, important for avoiding errors.
Typical FP dilution series:
| A | B | C | D |
|---|---|---|---|
| 1 | 100 of elution | 900 | 500 |
| 2 | 500 of prev dilution | 500 | " |
| 3 | " | " | " |
| 4 | " | " | " |
| 5 | " | " | " |
| 6 | " | " | " |
| 7 | " | " | " |
| 8 | " | " | " |
| 9 | " | " | " |
| 10 | " | " | " |
| 11 | " | " | " |
2. Fill standard clear 96-well plate
Typical arrangements:
Clear Plate: 200ul per well. 200ul per well.
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FP1 | row1 | A | dilution1 (neat) | dilution2 (1:2) | d3 | d4 | d5 | d6 | d7 | d8 | d9 | d10 | d11 | buffer |
| row2 | B | dilution1 (neat) | dilution2 (1:2) | d3 | d4 | d5 | d6 | d7 | d8 | d9 | d10 | d11 | buffer | |
| FP2 | C | |||||||||||||
| D | ||||||||||||||
| FP3 | E | |||||||||||||
| F | ||||||||||||||
| FP4 | G | |||||||||||||
| H |
Steps:
- Distribute 200ul buffer into column 12 with multipipette (Dispensing mode; 5ml; 200ul * 8; Asp 4, Disp 5).
- Add protein dilutions to plate with single channel manual pipette. Pipette tip use can be minimised if dispensing from the lowest concentration protein (dilution11) first.
3. Measure absorbance in clear plate - ECmax assay
Prewarm plate reader to temp used for cell growth curve assays to make sure the calibrations are valid for those temperatures - eg. 30oC.
Plate reader runs with clear plate:
| A | B | C | D | E | F |
|---|---|---|---|---|---|
| 1 | instrument1 | clear | none | A200-1000 | 2 min |
Only one scan is needed here. That is an absorbance scan between 200nm and 1000nm. On the Spark this is the maximum range you can do, and you don't need more anyway. The ECmax wavelength will vary, and the high wavelength is required for path length quantification.
Quantification of FP Fluorescence
The purpose of this step is to quantify the fluorescence of each FP dilution in the same plate that the E. coli growth curves will be carried out in .
[ Day 3 contd ]
Measure fluorescence in clear plate
Plate reader runs with clear plate (continued):
| A | B | C | D | E | F |
|---|---|---|---|---|---|
| 1 | instrument1 | clear | seal | FP-dependent fluorescence channel1 | 10min |
| 2 | instrument1 | clear | seal | FP-dependent fluorescence channel2 | 10min |
| 3 | instrument2 | clear | seal | FP-dependent fluorescence channel1 | 10min |
| 4 | instrument2 | clear | seal | FP-dependent fluorescence channel2 | 10min |
Fluorescence data should be gathered on a range of gains (I use 40-120), for one (or more) channels, as required. For eg. while mScarlet is best measured at emission channel 595/35, it can be useful to measure it in 620/20 as well, in cases where the 595/35 channel would pick up too much crosstalk from another FP (such as YFP).
How I name channels (filter sets):
Channels are named as a combination of the excitation, then emission filter, which are given names to indicate the region of the visible spectrum they cover, followed by numbers. eg. the standard GFP filter set is ‘green1green2’, which excites at wavelength 485nm (bandwidth 20nm) and measures emission at wavelength 535nm (bandwidth 25nm).
Protein storage
Proteins should be stored in the fridge protected from the light, which testing indicated kept proteins stable and active for up to 4 weeks. Long term storage and re-testing post freeze-thaw has not been tested.
Analysis - Calibration of Plate Reader
The aim of this protocol is that for a given FP, we can relate the number of FP molecules to the 'relative' fluorescence units observed in a given instrument, with a given filter set, and gain. The previous steps described how to purify FPs to produce calibrants and how to run the assays. The output data from such assays must now be analysed to obtain calibrations.
To assist with this process, we have developed an R package called FPCountR. The full details are on the GitHub page. In brief, functions in this package are provided for each analytical step. parser() functions parse plate reader data into idealised formats. get_concentration() functions calculate protein concentration from absorbance data. The generate_cfs() function obtains conversion factors using concentration and fluorescence data. Finally, process_plate() and calc_percell() functions extract data from microbial growth curves in units of molecules, and molecules/cell.

