Establishment of craniofacial exocrine gland organoid magnetic bioassembly platforms as aging multi-omic signatures
Teerapat Rodboon, Glauco Souza, Apiwat Mutirangura, Joao N. Ferreira
Lacrimal gland
Salivary gland
Xerophthalmia
Xerostomia
Aging
Cellular senescence
Organoids
Bioprinting
Abstract
In the last decade, relevant biotechnology advances took place in the biofabrication of craniofacial exocrine gland organoids mimicking lacrimal and salivary glands. Though certain challenges still remain not only due to the lack of protocols for organoid reproducibility but also towards the scarcity of methodologies for creating preclinical disease models with aging multi-omic signatures. Previously, our research group successful developed three-dimensional (3D) bioassembly technologies towards the generation of functional epithelial gland-like organoids via magnetic 3D bioprinting platforms (M3DB). To meet the needs of our aging Asian societies, a next step was taken to design consistent M3DB protocols for bioengineering organoid models with aging molecular and pathological features for lacrimal glands (LG) and salivary glands (SG). Herein, we established a feasible step-by-step protocol for producing both LG and SG organoids using M3DB platforms. Such a protocol provided reproducible preliminary outcomes resembling LG/SG organoids. Both acinar and ductal epithelial compartments were prominent (21 4.32% versus 42 6.72% of total cells, respectively), and could be clearly identified in these organoids. Meanwhile, these can be further developed into aging signature models by inducing cellular senescence via chemical mutagenesis. The generation of senescence-like organoids is our ultimate milestone aiming towards high throughput applications for drug screening and discovery and gene therapy investigations to reverse aging.
Before start
Steps
Mechanical and enzymatic primary cell dissociations from craniofacial exocrine glands
Gland dissection and mechanical tissue dissociation
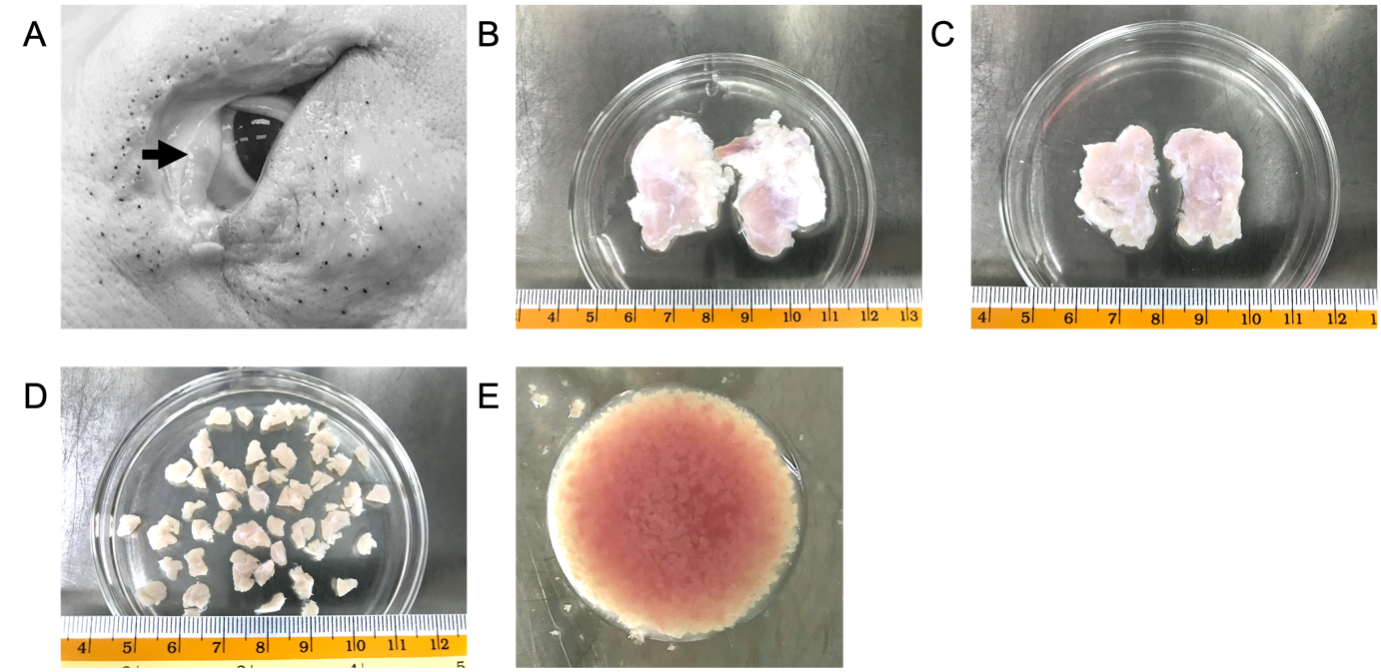
Lacrimal gland (LG) and salivary submandibular gland (SG) are dissociated from a 3 to 5 month-old porcine head. The head is carried inside an ice-containing box and delivered to the laboratory within 8h 0m 0s after animal sacrifice.
Mince the tissue using a surgical curved scissor into an apple sauce-like slurry paste (Figure 1D) and transfer the slurry into a 15 mL conical tube with a pre-wet 2.5 mL disposable Pasteur pipette.
Add 10mL of washing buffer and pipette up and down by using a pre-wet 10 mL serological pipette.
Let the tissue settle at the bottom of the tube by gravity for 0h 3m 0s and remove the supernatant using a pre-wet 10 mL serological pipette.
Repeat steps to for 3-5 times until the supernatant is clear.
Centrifuge the tissue slurry at 1000x g and carefully discard the supernatant.
Add 2mL (for 200 mg of tissue) of digestion buffer into a tissue fragment and gently mix well by vortexing. Make sure that no tissue fragment sticks on the lateral walls of the tube after vortexing.
Wrap the cap and neck of the tube with parafilm to minimize contamination.
Place the tube into a 37°C water beaker with magnetic stirring at 500 rpm and incubate for 0h 30m 0s (vortex the tube every 0h 15m 0s).
Refresh the enzymatic activity by repeating step to one more time.
The enzymatic dissociation activity is stopped by a dilution technique.
Clean the porcine head with sterilized water to remove any debris from the skin before disinfecting it with 0.5% (v/v) peracetic acid solution for 0h 15m 0s.
Add 8mL mL of washing buffer into the mixture and gently mix by pipetting for 3-5 times using a pre-wet serological pipette.
Centrifuge the mixture at 2000x g and discard the supernatant.
1.2.1Repeat step 1.2.12-1.2.13 for two more times.
Resuspend the cell pellet by adding 2mL of washing buffer and mix vigorously with a pre-wet 10 mL serological pipet.
Transfer the mixture by using the same pipette to the top of a mesh filter (100 µm pore size).
Wash the serological pipette and mesh filter with an additional 3mL washing buffer.
Collect the flow-through cell suspension.
To make a single cell suspension solution, gently aspirate the flow-through cell suspension by using a 29 G syringe. Then, gently pass the suspension through a 40 µm mesh strainer by pressing against the mesh on a circular motion.
Wash the syringe and mesh strainer with an additional 5mL washing buffer.
Centrifuge the cell suspension at 1000x g and discard the supernatant.
Gently wash with 1L of sterilized reverse osmosis (RO) water for 5 times and dry the skin by applying tissue paper.
Carefully aspirate the supernatant by a pre-wet P1000 pipette tip (~300 µL of buffer can be left on the pellet).
Add 2mL of culture media into the cell suspension and gently pipetting with a pre-wet P1000 pipette tip.
Assess the quality of the primary cells by determining the cell numbers and viability with the Trypan blue exclusion method and can confirm such counts with a
Equipment
| Value | Label |
|---|---|
| Countess 3 FL Automated Cell Counter | NAME |
| Automated Cell Counter | TYPE |
| Thermofisher scientific | BRAND |
| AMQAF2000 | SKU |
Cell plating and culture
One day before cell plating, thaw a vial of
For T75 flask coating, pipette 10µL of the BME into a 15 mL tube containing 5mL of cold serum-free
Place a vial of BME on ice during work to prevent untimely gelling.
Pipetting solution up and down with a 5mL serological pipette by being careful not to create air bubbles and then transfer the mixture into a T75 tissue culture flask.
Gently swirl the mixture to cover the entire growth surface area and incubate the flask at Room temperature for 1h 0m 0s.
Remove the mixture after incubation. The flask is ready for cell plating or can be kept in 4°C for plating on the following day.
For cell plating, pipette 1.0x105 cells in 10mL of expansion media into a BME-coated T75 culture flask.
Disinfect the skin around the eyes (for the LG) and at the angle of the mandible (for the SG) with 70% (v/v) ethanol followed by 1% (w/v)
Incubate cells at 37°C 5% CO2.
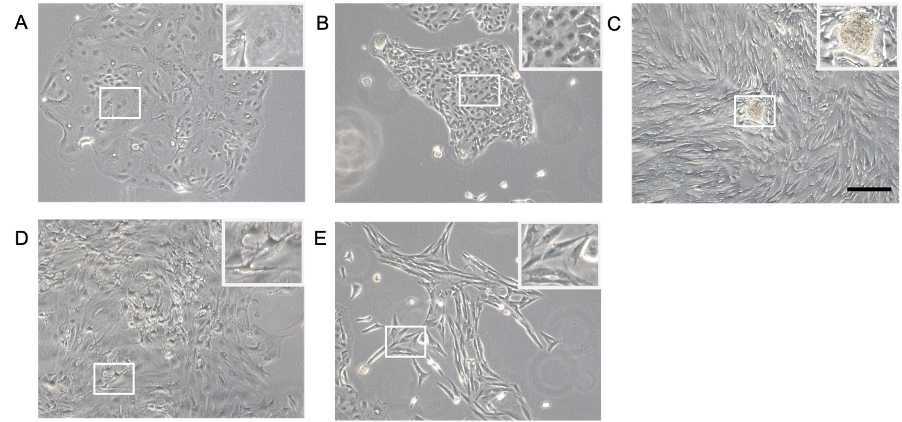
Observe the morphological heterogeneity (Figure 2) under a light microscope and replace the media every 2 days.
Cell passaging and epithelial enrichment and sorting
After the confluency of the monolayer cells reached 70%-80%, remove the old expansion media from a culture flask with a 10 mL serological pipette.
Transfer 10mL of 1XPBS into the flask and incubate for 0h 1m 0s.
Discard the solution from the cells before pipetting 1mL of
Incubate with the enzyme solution at 37°C for 0h 15m 0s. To enhance the cell dissociation process, remove the flask from the incubator to swirl or shake every 0h 5m 0s.
Observe the single cell dissociation phenomena under a light microscope.
Stop the enzymatic reaction using a dilution technique: use a 10 mL serological pipette to transfer 9mL of basal medium into a flask and resuspend the suspension by pipetting up and down for 3-5 times.
Transfer the suspension into a 15 mL conical tube and pellet the cells by centrifugation at 1000x g.
Discard the supernatant and resuspend the pellet by using 5mL of expansion media.
Collect the glands in a 50 cm Petri dish (Figure 1B) and disinfect with 70% (v/v) ethanol. To protect the dry out of tissue, covering the gland with 2mL of collection media.
Assess the cell number and viability by using Trypan blue exclusion method and confirm the cell count with
Equipment
| Value | Label |
|---|---|
| Countess 3 FL Automated Cell Counter | NAME |
| Automated Cell Counter | TYPE |
| Thermofisher scientific | BRAND |
| AMQAF2000 | SKU |
Pipette 1.0x105 cells in 10mL of expansion medium into a new BME-coated T75 tissue culture flask and incubate the cells at 37°C 5%CO2.
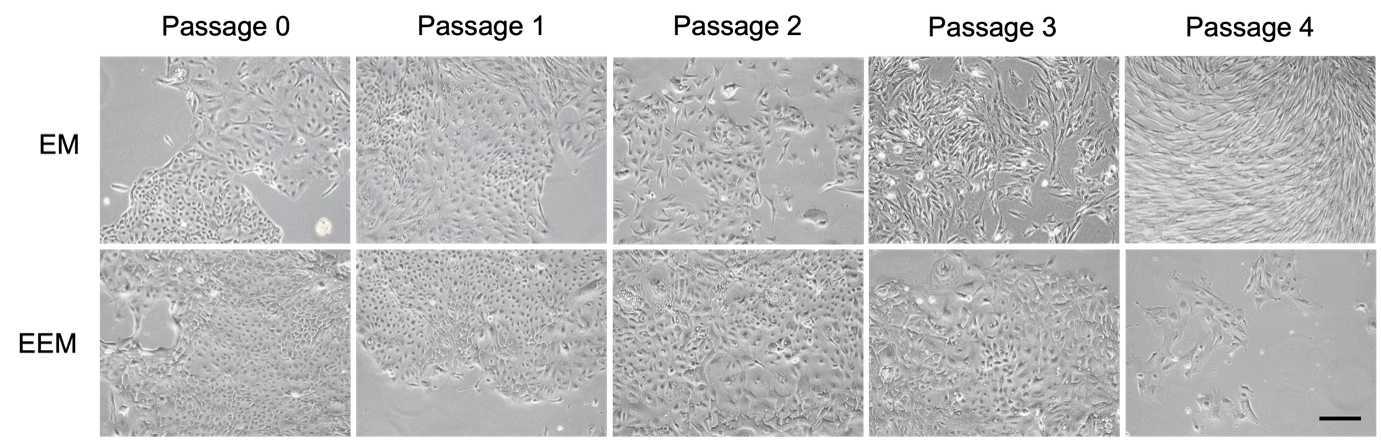
To passage the cells or perform epithelial cell enrichment and/or sorting. Replace the old expansion medium with 10mL of fresh EM or EEM at day 2 and replace the medium every 48h 0m 0s until cells reach the desired confluency.
Remove peripheral fat and connective tissue by using precision forceps and scissors (Figure 1C) before sectioning the tissue with a scalpel into 0.5 cm3 to 1 cm3 pieces (Figure 1D).
Transfer tissue into a 50 mL collection tube and wash with 20mL collection media for 3-5 times or until the solution is clear.
Keep the tissue in 20mL collection media at 4°C to7°C for 6h 0m 0s to8h 0m 0s.
Primary cell extraction and isolation
Transfer 200mg of gland tissue into a glass spot plate and add 500µL of collection buffer to prevent tissue to dry out.
Aging Organoid Establishment with Magnetic Bioassembly
Cell magnetic bioassembly and senescence induction
2.1.1Before magnetization, dissociate the cells at the monolayer stage at the confluency of 70-80% by following the previous steps in section 1 .
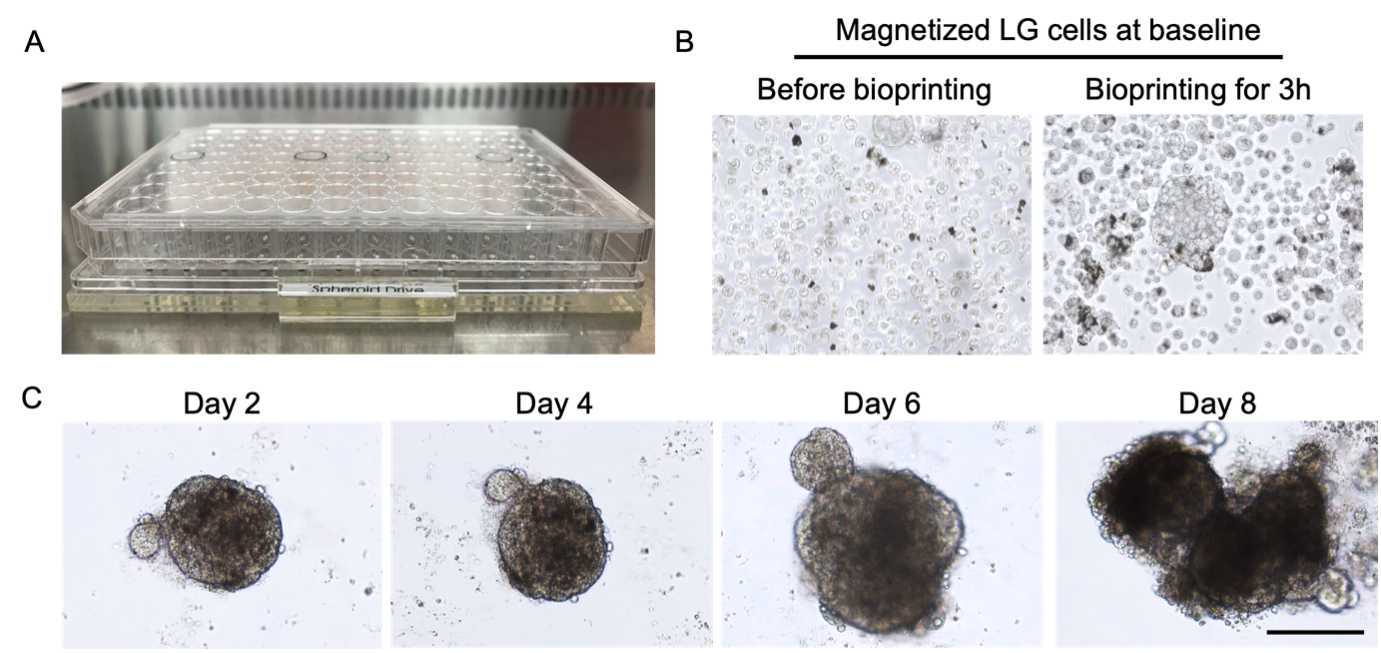
Place the ultra-low attachment 96 well plate on top of a 96-well spheroid magnetic drive prior to bioassembly and bioprinting (Figure 4A).
Transfer 150µL of cell suspension to each well of the plate. To prevent aggregation of the magnetized cells, gently hand shake the tube during pipetting.
Seal the border of the plate with 100µL of sterilized water to minimize the evaporation of media and incubate the plate in 37°C , 5% CO2 with humidity for 3h 0m 0s and observe cell morphology (Figure 4B).
Remove the magnetic drive from the bottom of the plate and incubate the plate further in 37°C , 5% CO2 with humidity for 192h 0m 0s with medium replacement every 48h 0m 0s (Figure 4C).
To induce cellular senescence, treat the organoids with 10millimolar (mM) -100millimolar (mM) of 24h 0m 0s.
Then, resuspend the cell pellet with 1mL of epithelial enrichment media (EEM).
Determine cell numbers and viability by Trypan blue exclusion method and confirm the count with a
Equipment
| Value | Label |
|---|---|
| Countess 3 FL Automated Cell Counter | NAME |
| Automated Cell Counter | TYPE |
| Thermofisher scientific | BRAND |
| AMQAF2000 | SKU |
.
To fabricate 20 organoids, prepare 420µL μL of the cells at a density 1.0x106 cells/mL in a 50 mm dish by adjusting with EEM.
Add 42µL of the
Incubate the suspension for 2h 0m 0s in a 37°C incubator, 5% CO2. To ensure proper mixing during incubation, shake the tube on an orbital shaker at 250rpm.
After incubation, centrifuge the cell-MNP solution at 800x g and remove the supernatant by pipetting.
Gently tap the cell pellet to resuspend the single cells and adjust the cell concentration to 1.33x105 cells/mL by adding 2730µL of expansion media.
Pipette the mixture up and down with a P1000 pipette tip to ensure a single cell suspension.
Secretory LG/SG organoid validation
3.1 Immunofluorescent staining of acinar and ductal epithelial compartments
Use a magnetic holder to hold the organoid at the bottom of each well in the 96 well plate and remove all media from the organoids. Avoid sucking up the organoids and shear them through the P200 pipette tip (always use normal uncut tip for this step).
After incubation, remove the blocking buffer from organoids with a P200 pipette.
Add 100µL solution of primary antibodies (against protein markers of acinar and ductal epithelial cells) into the organoids and incubate for at least 3h 0m 0s at Room temperature or 4°C in a humidified chamber with 400rpm orbital shaker.
After incubation, remove the solution from the organoids with a P200 pipette tip and wash the excess of antibody solution with 200µL of 0.1% (v/v) Tween-20 in 1X PBS for 0h 20m 0s with 400rpm orbital shaker, at least three times.
Add 100µL of a solution with secondary antibodies into the organoids (specific to the host species of the previously used primary antibodies) and incubate at room temperature for 1h 0m 0s with a protection against photobleaching.
After incubation, remove all solutions with secondary antibodies with a P200 pipette tip and rinse with 200µL of a washing buffer solution containing 0.5% (v/v) Tween-20 in 1X PBS for 0h 20m 0s with 400rpm orbital shaker, at least three times.
Replace the solution with 100µL of nuclear stain solution (10% (v/v) Hoechst 33342 in 1X PBS) and incubate at Room temperaturewith 400rpm orbital shaker for 1h 0m 0s.
Remove the nuclear stain solution and observe the labeled organoids under a fluorescence microscope before mounting them on a regular glass slide with a
Antibodies used
| A | B | C | D |
|---|---|---|---|
| Rabbit monoclonal anti-AQP-5 IgG | Abcam | AB92320 | 1:100 |
| Rabbit monoclonal anti-KRT14 IgG | Abcam | AB181595 | 1:100 |
| Rabbit monoclonal anti-KRT19 igG | Novus Biologicals | NBP142238 | 1:100 |
| Alexa Flour488 goat anti-rabbit IgG | Abcam | AB150077 | 1:200 |
| Alexa Flour488 goat anti-mouse IgG | Abccam | 150113 | 1:200 |
Antibodies
Solutions/media used
| A | B | C |
|---|---|---|
| 1XPBS | Not applicable | 90 |
| 1XPBSPenicillin/Streptomycin (100%) | 10% v/v | 10 |
| Total | 100 |
| A | B | C |
|---|---|---|
| 1XPBS | Not applicable | 86.67 |
| 1XPBSPenicillin/Streptomycin (100%) | 10%v/v | 10 |
| Bovine resum albumin (30%) | 1%v/v | 3.33 |
| Total | 100 |
| A | B | C |
|---|---|---|
| 1XPBS | Not applicable | 1.763 |
| 1XPBSPenicillin/Streptomycin (100%) | 1%v/v | 0.020 |
| Bovine resum albumin (30%) | 1%v/v | 0.067 |
| Calcium chloride solution (50 mM) | 1.25 mM | 0.050 |
| Collagenase II (40 mg/mL) | 1 mg/mL | 0.050 |
| Hyaluronidase | 1 mg/mL | 0.050 |
| Total | 2 |
| A | B | C |
|---|---|---|
| DMEM/F12 | Not applicable | 98 |
| L-Glutamine (100 mM) | 1 mM | 1 |
| Penicillin/Streptomycin (100%) | 1% v/v | 1 |
| Total | 100 |
| A | B | C |
|---|---|---|
| Basal media | Not applicable | 94.85 |
| Fetal bovine serum (100%) | 5%v/v | 5 |
| EGF (20 µg/mL) | 20 ng/mL | 0.1 |
| Total | 100 |
| A | B | C |
|---|---|---|
| Define Keratinocyte SFM | Not applicable | 99.80 |
| EGF (20 µg/mL) | 20 ng/mL | 0.1 |
| FGF-10 (100 µg/mL) | 50 ng/mL | 0.05 |
| FGF-7 (100 µg/mL) | 50 ng/mL | 0.05 |
| Total | 100 |
| A | B | C |
|---|---|---|
| 1XPBS | Not applicable | 0.633 |
| Fetal bovine serum (100%) | 5%v/v | 0.1 |
| Horse serum (100%) | 10%v/v | 0.167 |
| 1% v/v Tween 20 | 0.1%v/v | 0.1 |
| Total | 1 |
| A | B | C |
|---|---|---|
| 1XPBS | Not applicable | 900 |
| Hoechst 33342 (100%) | 10%v/v | 100 |
| Total | 1 |
Gently wash the organoids with 200µL of 1X PBS and discard all solution.
Fix the organoids by adding 100µL of 4Mass / % volume 0h 30m 0s with a 400rpm orbital swirling.
Use a magnetic holder to hold the organoid in each well of 96 well plate and remove all solutions (4% PFA and later 1X PBS) from the organoids.
Gently wash the organoids with 200µL of 1X PBS and discard all solution for three times.
For immunofluorescent labeling, remove residual 1X PBS with a P200 pipette tip as much as possible.
Permeabilize the organoids with 200µL of 0.1% (v/v) Triton X for 0h 20m 0s with 400rpm orbital swirling and then remove all solution with a P200 pipette tip.
Wash the organoids with 200µL of 1X PBS and remove the solution with a P200 pipette tip for at least three times.
Add 200µL of blocking buffer into the center of organoids and incubate for 6 hours at room temperature or 1h 0m 0s - 2h 0m 0s at Room temperature followed by at 4°C in a humidified chamber with a 400rpm orbital shaker.

