Differentiation of iPSCs with the hNIL construct into motor neurons protocol
Maria Sckaff, Kenneth Wu, Hana Ghanim, Aradhana Sachdev, Gokul N Ramadoss, Carissa M. Feliciano, Luke M. Judge, Bruce Conklin, Claire D Clelland
Abstract
This protocol describes the differentiation of induced pluripotent stem cells (iPSCs) into motor neurons using the hNIL transgenic factors in a CLYBL safe harbor site.
Attachments
Steps
Maintenance and Preparation of the iPSCs for the hNIL differentiation into motor neurons: Maintaining iPSCs
Culture iPSCs for at least 2-3 passages after initial hNIL transfection or from frozen stocks before starting a motor neuron differentiation.
Briefly, we maintain iPSCs on Matrigel-coated plastic culture dishes (Growth Factor Reduced Matrigel diluted in 50mL of KnockOut DMEM to a concentration of 80) with mTeSR™ Plus medium.
Change media every other day, and passage every 4-5 days with ReLeSR™ (see below).
Add ROCK1 inhibitor at 10micromolar (µM) in media to freshly passaged iPSCs for 1 day to limit spontaneous differentiation.
Maintenance and Preparation of the iPSCs for the hNIL differentiation into motor neurons: Passaging iPSCs
Passage iPSCs as clumps (for routine expansion of iPSC cultures) or single cells (for differentiation).
For routine expansion, wash the well with DPBS -Ca/-Mg (1-2 mL/well*) then add 1mL per well* of ReLeSR™ for 0h 1m 0s.
Aspirate most of the ReLeSR™, but do not overdry, and incubate the wells at Room temperature for 3-4 minutes.
Add fresh mTeSR™ Plus media to the well (1mL/well*) and pipette gently to detach the iPSCs and break colonies into small clumps.
A confluent well can be split 1:6 to 1:20 depending on the desired confluency and rate of growth of the iPSC line.
If desired, before replating, cells can be centrifuged at 800rpm to pellet clumps and remove any single cells (which remain in suspension).
For differentiation (or anytime cell counting is needed), wash the well with DPBS -Ca/-Mg (1-2 mL/well*) then add 0.5mL* of Accutase®.
Incubate the plate for 0h 5m 0s at 37°C.
After the incubation, tap the plate to fully detach the iPSCs, then add 0.5mL* of KnockOut DMEM with 20% FBS (alternately, 2.5 mL/well* of DPBS +Ca/+Mg can be used in place of DMEM/FBS).
Transfer the cells and solution to a conical vial and centrifuge for 800rpm to pellet the cells.
Aspirate the supernatant carefully as to not disturb the cell pellet, and resuspend the cells in mTeSR™ Plus with ROCK1 inhibitor at 10micromolar (µM).
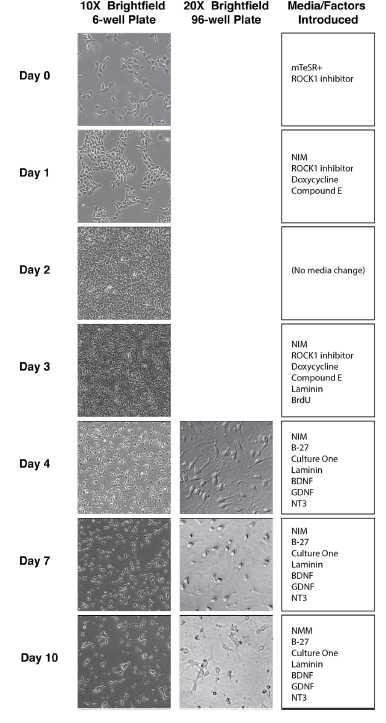
hNIL differentiation into motor neurons: Day 0
Coat the receiving vessel with Matrigel at least 0h 30m 0s before starting Day 0 (but no more than 24-36 hours prior to starting) and incubate at 37°C.
Bring the mTeSR™ Plus with ROCK1 inhibitor (final concentration of 10micromolar (µM) in well, 1000X dilution from 10millimolar (mM) stock) and the Accutase® to 37Room temperature.
Aspirate the spent media and wash the wells with cells once with 1X DPBS -Ca/-Mg.
Add appropriate volume of Accutase® per well.
Incubate the plate for 0h 5m 0s at 37°C.
Remove plate from the incubator and tap the plate to release the cells.
Quench the Accutase® using five times the volume DPBS +Ca/+Mg (if you used 0.5mL of Accutase®, quench with 2.5mL of DPBS).
Transfer the cells to a conical vial.
Centrifuge the vial at 800rpm.
Aspirate the Accutase® with DPBS, being careful not to disturb the cell pellet at the bottom of the vial.
Resuspend the cells with an appropriate volume of mTeSR™ Plus with ROCK1 inhibitor (final concentration of 10micromolar (µM) in well, 1000X dilution from 10millimolar (mM) stock). Aim for roughly 1mL of mTeSR™ Plus with ROCK1 inhibitor per 1 million cells.
Pipette up and down to mix well and produce an evenly distributed solution.
Count the number of cells using a Countess II (any preferred equivalent way of counting cells is also appropriate).
Aspirate the Matrigel from the receiving vessel.
Add appropriate volume of media (the mTeSR™ Plus with ROCK1 inhibitor at 10millimolar (mM)) to the receiving vessel.
Add volume of cell mixture appropriate for the number of cells desired in the well. For a 10cm dish, we have found 1-2 million iPSCs at day 0 produces 3-5 million cells at day 3.
Place the receiving vessels (which now has your cells) back in the incubator at 37°C.
Shake the plate in all four directions to ensure the cells are evenly distributed in the well.
hNIL differentiation into motor neurons: Day 1
Prepare the neural induction medium (NIM) as follows:
| A | B |
|---|---|
| DMEM/F12 | 97 mL |
| N-2 | 1 mL |
| NEAA | 1 mL |
| GlutaMAX | 1 mL |
Calculate the volume of freshly prepared NIM needed to perform a full media change (see Table 2) and aliquot in a vial of appropriate size.
| A | B | C | D | E |
|---|---|---|---|---|
| Cell culture vessel | Surface area (cm2) | Media volume per well | Day 3 Seeding density for ICC applications | Day 3 Seeding density for protein, RNA, DNA extraction |
| 96-well plate | 0.32 | 150 µL | 20-25k cells | - |
| 24-well plate | 1.9 | 500 µL | 80-100k cells | - |
| 12-well plate | 3.5 | 1 mL | 2 x 105 cells | - |
| 6-well plate | 9.6 | 2 mL | 0.5 x 106 cells | 1 x 106 cells |
| 10-cm dish | 56.7 | 10 mL | - | 5 x 106 cells |
| 15-cm dish | 145 | 30 mL | - | 15 x 106 cells |
Table 2: Number of cells and volume of media matrix needed on the day-3 replating in the differentiation of hNIL iPSCs into motor neurons based on the size and format of the cell culture vessel used, as well as the application. Modification from the Kampmann’s Lab protocol3.
Add the appropriate factors for day 1 to the NIM aliquot you aliquoted in step 35:
| A | B | C | D |
|---|---|---|---|
| Factor | Final Concentration | Stock | Dilution |
| ROCK1 inhibitor | 10 μM | 10 mM | 1:1,000 |
| Doxycycline | 2 μg/mL | 2 mg/mL | 1:1,000 |
| Compound E | 0.2 μM | 2 mM | 1:10,000 |
Aspirate the spent media from the wells with cells.
Add the appropriate volume of media to each well.
Place the plate with cells in fresh media back in the incubator at 37°C.
hNIL differentiation into motor neurons: Day 2
Select the receiving cell-culture vessel. This is the plate on which your neurons will grow permanently (see recommendations below).
Coat the wells which will be receiving cells on day 3 with the appropriate volume of 1X PDL at 0.1mg/mL (see Table 1).
| A | B | C | D |
|---|---|---|---|
| Cell culture vessel | Matrigel, PDL, and laminin in KO DMEM volume per well | DPBS volume for washes per well | Accutase® volume per well |
| 96-well plate | 120 µL | 150 µL | 50 µL |
| 24-well plate | 250 µL | 400 µL | 125 µL |
| 12-well plate | 500 µL | 1 mL | 250 µL |
| 6-well plate | 1 mL | 2 mL | 500 µL |
| 10-cm dish | 4 mL | 6 mL | 3 mL |
| 15-cm dish | 12 mL | 18 mL | 9 mL |
Table 1: Volumes of coating solutions (Matrigel, PDL, and laminin in KO DMEM), DPBS, and Accutase® for each cell culture vessel.
Place the coated plate back in the incubator at 37°C.
hNIL differentiation into motor neurons: Day 3; Part 1
Wash the receiving wells.
Wash the receiving wells with 1X DPBS. (1/2)
Wash the receiving wells with 1X DPBS. (2/2)
Let receiving wells dry in the laminar flow hood for 20-30 minutes.
Coat receiving cell-culture vessel with the appropriate volume of NIM with laminin mouse protein (at a concentration of 15) for 2-5 hours at 37°C (see Table 1 for volume)
| A | B | C | D |
|---|---|---|---|
| Cell culture vessel | Matrigel, PDL, and laminin in KO DMEM volume per well | DPBS volume for washes per well | Accutase® volume per well |
| 96-well plate | 120 µL | 150 µL | 50 µL |
| 24-well plate | 250 µL | 400 µL | 125 µL |
| 12-well plate | 500 µL | 1 mL | 250 µL |
| 6-well plate | 1 mL | 2 mL | 500 µL |
| 10-cm dish | 4 mL | 6 mL | 3 mL |
| 15-cm dish | 12 mL | 18 mL | 9 mL |
hNIL differentiation into motor neurons: Day 3; Part 2
Prepare quenching media DMEM/F12 with 20% FBS at an appropriate volume. Per well in a 6- well plate, 0.5mL of quenching media will be needed.
Calculating the volume of day 3 NIM media needed
| A | B | C | D | E |
|---|---|---|---|---|
| Cell culture vessel | Surface area (cm2) | Media volume per well | Day 3 Seeding density for ICC applications | Day 3 Seeding density for protein, RNA, DNA extraction |
| 96-well plate | 0.32 | 150 µL | 20-25k cells | - |
| 24-well plate | 1.9 | 500 µL | 80-100k cells | - |
| 12-well plate | 3.5 | 1 mL | 2 x 105 cells | - |
| 6-well plate | 9.6 | 2 mL | 0.5 x 106 cells | 1 x 106 cells |
| 10-cm dish | 56.7 | 10 mL | - | 5 x 106 cells |
| 15-cm dish | 145 | 30 mL | - | 15 x 106 cells |
Table 2: Number of cells and volume of media matrix needed on the day-3 replating in the differentiation of hNIL iPSCs into motor neurons based on the size and format of the cell culture vessel used, as well as the application. Modification from the Kampmann’s Lab protocol3.
Preparing day 3 NIM media:
| A | B | C | D |
|---|---|---|---|
| Factor | Final Concentration | Stock | Dilution |
| ROCK1 inhibitor | 10 μM | 10 mM | 1:1,000 |
| Doxycycline | 2 μg/mL | 2 mg/mL | 1:1,000 |
| Compound E | 0.2 μM | 2 mM | 1:10,000 |
| Laminin | 1 μg/mL | 1 mg/mL | 1:1,000 |
| BrdU | 40 μM | 40 mM | 1:1,000 |
Aspirate the spent media and wash the wells with cells once with 1X DPBS -Ca/-Mg.
Add appropriate volume of Accutase® per well (0.5mL/well in a 6-well plate).
Incubate the plate for 0h 5m 0s at 37°C.
Remove plate from the incubator and tap the plate to release the cells.
Quench the Accutase® with DMEM/F12 + 20% FBS, at a volume equal to the volume of Accutase® per well (if you used 0.5mL of Accutase®, quench Accutase® with 0.5mL of DMEM/F12 + 20% FBS). DPBS +Ca/+Mg is also suitable.
Pipette the solution gently around the well thoroughly to ensure cells have dissociated from the well bottom.
Move the cells from their wells to a 15-mL vial (or vial of appropriate volume depending on how much volume of cells you have).
Centrifuge the vial at 800rpm.
Aspirate the Accutase® with DMEM/F12 + 20% FBS, being careful not to disturb the cell pellet at the bottom of the vial.
Resuspend the cells with an appropriate volume of day 3 NIM media (about 1 mL per 1 million cells in the vial).
Count the cells using a Countess™ (or preferred counting method) using the same parameters from Day 0.
Aspirate the NIM with 15 laminin from each receiving well.
Plate appropriate number of cells per well using the appropriate media volume per well depending on your receiving cell culture vessel.
Sample calculation and coating advice for uniform seeding of cells onto receiving vessel:
Calculate the volume of day 3 NIM media needed per well.
Multiply that volume by the desired concentration of cells per well (e.g., 20k cells/well or 1 x 106 cells/well).
Divide that number by the concentration found by counting using the Countess™ (e.g. 2 x 106 cells/well).
The resulting number is the volume you need from the cell solution you resuspended in day 3 NIM media in step 59.
Add that volume to your final desired volume of day 3 NIM media (e.g., 4mL if seeding onto 2 wells of a 6-well plate).
Mix well to ensure a uniform distribution of cells.
Seed appropriate media volume (which now has cells as well) onto the receiving cell culture vessel.
Carefully place the plate with cells back in the incubator at 37°C, rocking the plate briefly after placing the plate back, such that the cells are uniformly distributed onto the well.
hNIL differentiation into motor neurons: Day 4
Prepare the day 4 NIM media for performing a full-media change
| A | B | C | D |
|---|---|---|---|
| Factor | Final Concentration | Stock | Dilution |
| B-27 Supplement | 50x | 01:50 | |
| Culture One Supplement | 100x | 1:100 | |
| Laminin | 1 μg/mL | 1 mg/mL | 1:1,000 |
| BDNF | 20 ng/mL | 20 μg/mL | 1:1,000 |
| GDNF | 20 ng/mL | 20 μg/mL | 1:1,000 |
| NT3 | 20 ng/mL | 20 μg/mL | 1:1,000 |
Aspirate the media slowly from the wells, using a pipette, discarding into a reservoir.
Add the fresh media slowly at a direction normal to the well side wall .
Place the plate back in the incubator at 37°C.
hNIL differentiation into motor neurons: Day 7
The risk of lifting only increases as the motor neurons age and mature on the well bottom.
Prepare the day 7 NIM media for performing a half-media change.
| A | B | C | D |
|---|---|---|---|
| Factor | Final Concentration | Stock | Dilution |
| B-27 Supplement | 50x | 1:50 | |
| Culture One Supplement | 100x | 1:100 | |
| Laminin | 500 ng/mL | 1 mg/mL | 1:1,000 |
| BDNF | 20 ng/mL | 20 μg/mL | 1:500 |
| GDNF | 20 ng/mL | 20 μg/mL | 1:500 |
| NT3 | 20 ng/mL | 20 μg/mL | 1:500 |
Aspirate the media slowly from the wells, using a pipette, discarding into a reservoir.
Add the fresh media slowly in at a direction normal to the well side wall (which is easier by tilting the plate about 60-degrees towards you).
Place the plate of motor neurons back in the incubator at 37°C.
hNIL differentiation into motor neurons: Day 10 on
Prepare Neurobasal Medium ( NMM ) as follows:
| A | B |
|---|---|
| Neurobasal | 97 mL |
| N-2 | 1 mL |
| NEAA | 1 mL |
| GlutaMAX | 1 mL |
Prepare the day 10 NMM media for performing a half-media change.
| A | B | C | D |
|---|---|---|---|
| Factor | Final Concentration | Stock | Dilution |
| B-27 Supplement | 50x | 1:50 | |
| Culture One Supplement | 100x | 1:100 | |
| Laminin | 500 ng/mL | 1 mg/mL | 1:1,000 |
| BDNF | 20 ng/mL | 20 μg/mL | 1:500 |
| GDNF | 20 ng/mL | 20 μg/mL | 1:500 |
| NT3 | 20 ng/mL | 20 μg/mL | 1:500 |
Aspirate the media slowly from the wells, using a pipette, discarding into a reservoir.
Add the fresh media slowly in at a direction normal to the well side wall (which is easier by tilting the plate about 60-degrees towards you).
Place the plate of motor neurons back in the incubator at 37°C.
Perform a half-media change with NMM and factors every 7 days for up to 9 weeks or more (if using the plate for imaging) or every 4 days (if using the plate for protein, RNA, or DNA extraction).