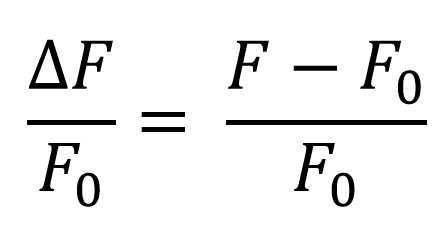Development of a simple and versatile in vitro method for production, stimulation, and analysis of bioengineered muscle
Karen Wells-Cembrano, Júlia Sala, Jose Antonio del Rio
Bioengineered muscle
skeletal muscle
in vitro model
electrical stimulation
optogenetics
acetylcholine
muscle stimulation
hydrogels
biomaterials
differentiation
Matrigel
Fibrinogen
Velcro
3D model
ChR2
Fluo-4 AM
ImageJ
Abstract
In recent years, 3D in vitro modeling of human skeletal muscle has emerged as a subject of increasing interest, due to its applicability in basic studies or screening platforms. These models strive to recapitulate key features of muscle architecture and function, such as cell alignment, maturation, and contractility in response to stimuli. To this end, it is required to culture cells in biomimetic hydrogels suspended between two anchors. Currently available protocols are often complex to produce, have a high rate of breakage, or are not adapted to imaging and stimulation. Therefore, we sought to develop a simplified and reliable protocol, which still enabled versatility in the study of muscle function. In our method, we have used human immortalized myoblasts cultured in a hydrogel composed of MatrigelTM and fibrinogen, to create muscle strips suspended between two VELCROTM anchors. The resulting muscle constructs show a differentiated phenotype and contractile activity in response to electrical, chemical and optical stimulation. This activity is analyzed by two alternative methods, namely contraction analysis and calcium analysis with Fluo-4 AM. In all, our protocol provides an optimized version of previously published methods, enabling individual imaging of muscle bundles and straightforward analysis of muscle response with standard image analysis software. Although limited by the lack of co-culture with motor neurons, this system provides a start-to-finish guide on how to produce, validate, stimulate, and analyze bioengineered muscle. This ensures that the system can be quickly established by researchers with varying degrees of expertise, while maintaining reliability and similarity to native muscle.
Before start
Steps
Making muscle culture devices
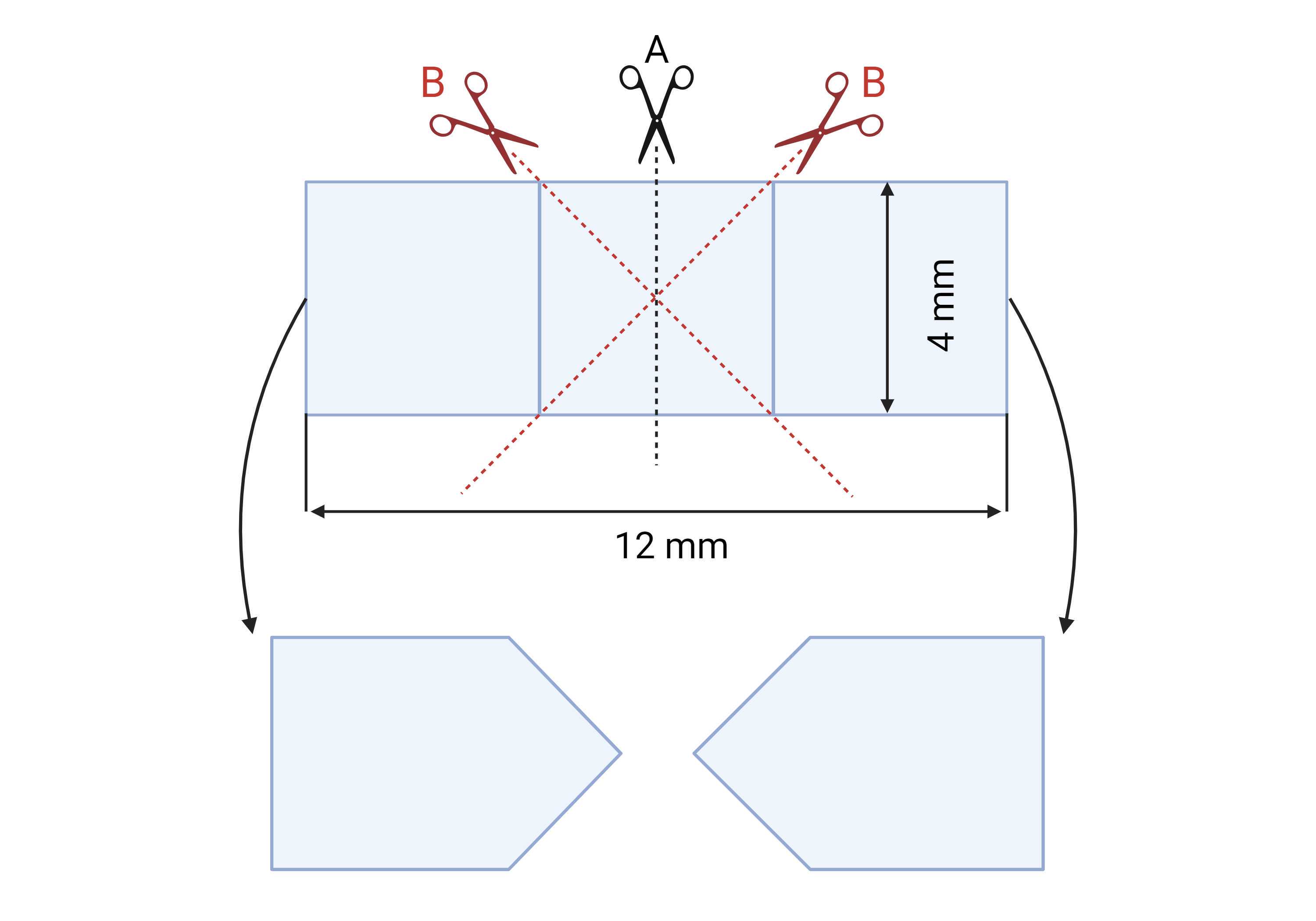
Prepare VELCRO ® pieces. Only use white velcro without adhesive backing. Using only the "loop" (soft) part of the VELCRO®, cut arrow-shaped velcro pieces according to directions in Figure 1 . Use a pencil (not a marker) to divide the VELCRO® into 12 x 4 mm strips, as shown in Figure 1 . Then, cut these strips in half ( Figure 1A ), which can be easily done by folding the strip and cutting along the fold. Next, cut the arrowheads as shown in Figure 1B .
C2C12ChR2 NO RECORDINGS
Prepare PDMS glue by mixing 1h 0m 0s to completely remove bubbles.
c21c12chr2 recording
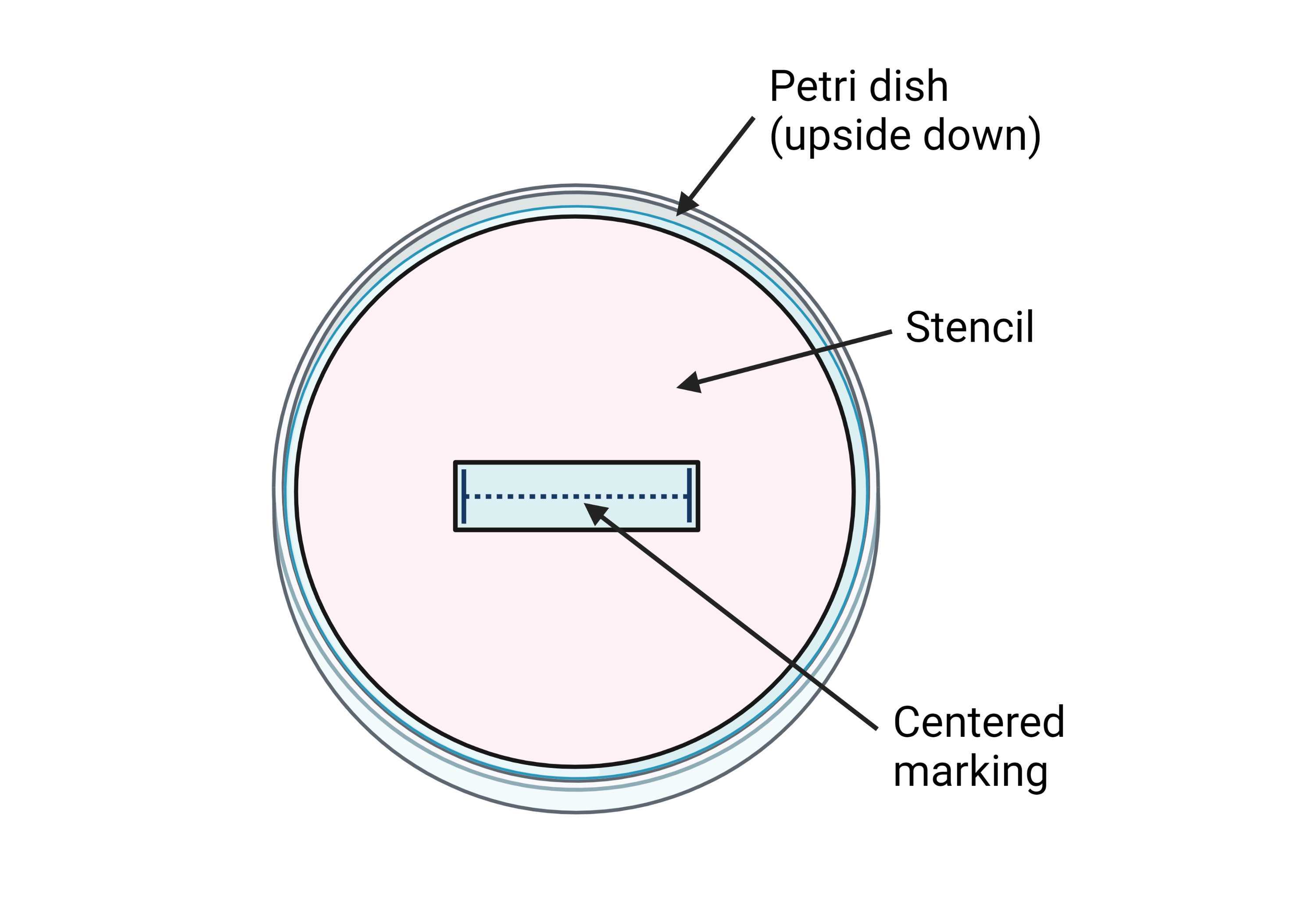
Mark a line of 1.2 cm in the center of each 35 mm ∅ Petri dish. In order to obtain consistently centered markings, it is recommended to make a small stencil out of paper or plastic ( Figure 2 ).
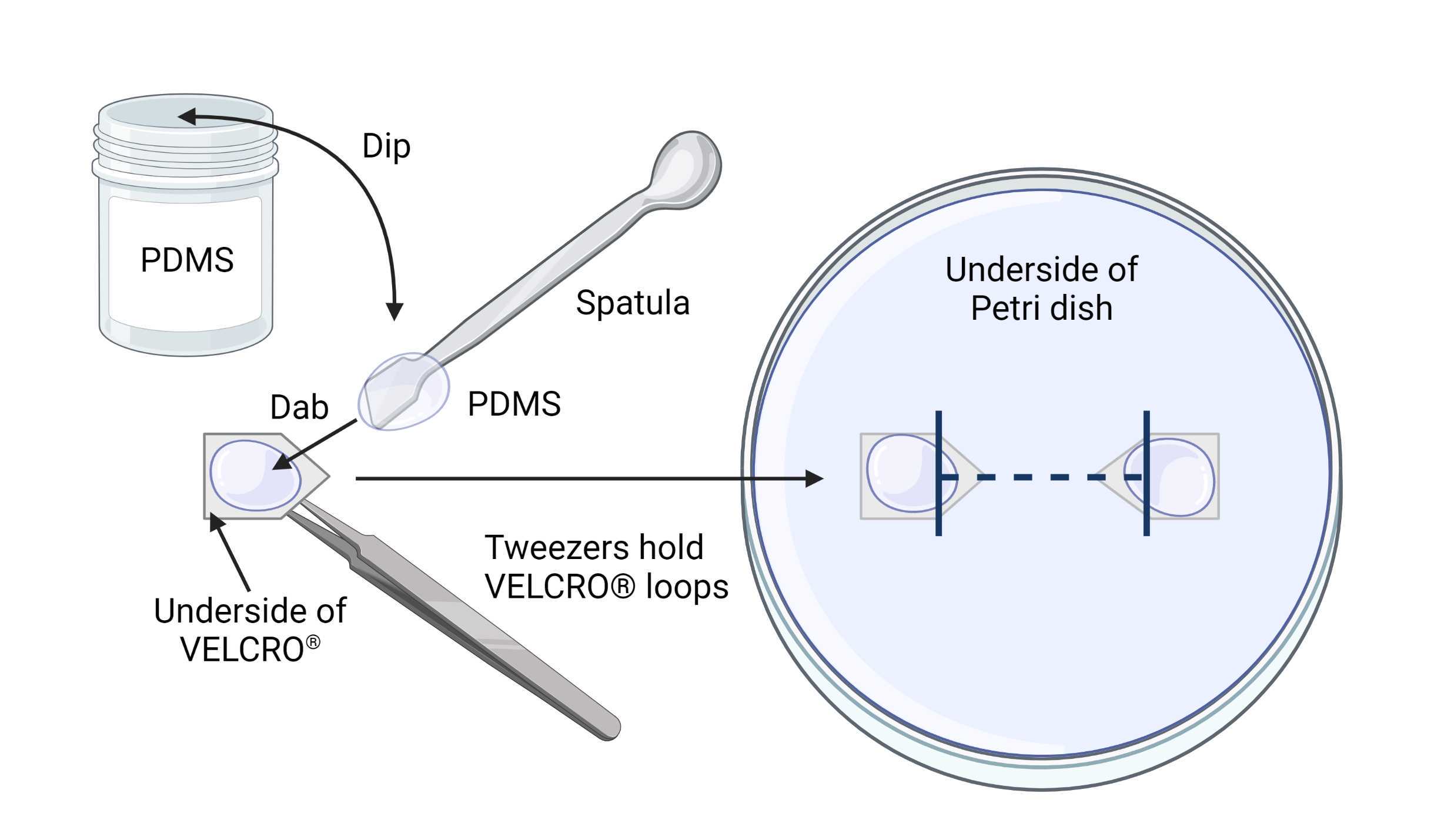
Attach VELCRO ® anchors to the Petri dishes, using the marked line as a guide. Use a small spatula to dab glue onto the underside of the VELCRO® pieces, and then place them on the lines using tweezers, as shown in Figure 3 . Make sure that the tips of the anchors are aligned, and the ends of the markings are aligned to the start of the arrowhead region of the anchors.
Once the VELCRO® pieces are attached, leave the plates at Room temperature to cure the PDMS glue . Then, store them until further use.
Preparation of cells and materials
Defrost myoblast cell line in a T75 flask and culture cells in their recommended maintenance medium. Human myoblasts must be passaged at a maximum of 60-70% confluence to avoid differentiation induced by cell-cell contacts.
For maintenance of human myoblasts, we use 1h 0m 0s before use.
Amplify cells to the needed amount. Take into account that around 3·106 cells are needed for each muscle bundle 6 cells are needed for each muscle bundle. If many replicates are to be seeded, a large number of flasks and high volume of cell culture medium may be necessary depending on the yield for each cell line. In the case of human myoblasts, cells are sometimes larger than other common cell lines, which can make it more difficult to obtain high yields of cells while keeping confluence under 70%.
Prepare a thrombin stock solution according to manufacturer's instructions, using
The day before the seeding, defrost the appropriate amount of 2h 0m 0s. It is recommended to start defrosting in the afternoon, and to change the ice the next morning.
The day of the seeding, prepare a stock solution of fibrinogen at 8mg/mL .
First, calculate the necessary amount of fibrinogen solution needed for your experiment, taking into account that 150µL are needed for each muscle bundle. Prepare 1.5 times the necessary volume.
Layer fibrinogen over warm (sterile) phosphate-buffered saline (PBS), leave it in the 37°C water bath to fully dissolve (DO NOT VORTEX), and filter through a 0.22 μm filter. Keep the solution at 4°C until use.
On the day of seeding, sterilize the necessary amount of Petri dishes according to the following steps.
Place the Petri dishes on larger (150 mm Ø or similar) Petri dishes, for ease of manipulation ( Figure 4 ). Add 70% ethanol to each dish, up to the brim, and leave for 0h 15m 0s .
After UV sterilization, turn the hood back on, and remove the PBS by aspiration with a glass Pasteur pipette and vacuum. Make sure to aspirate all of the liquid from the VELCRO® anchors, which are quite absorbent. Then, leave the plates in the incubator for approximately 1h 0m 0s to dry the plates completely.
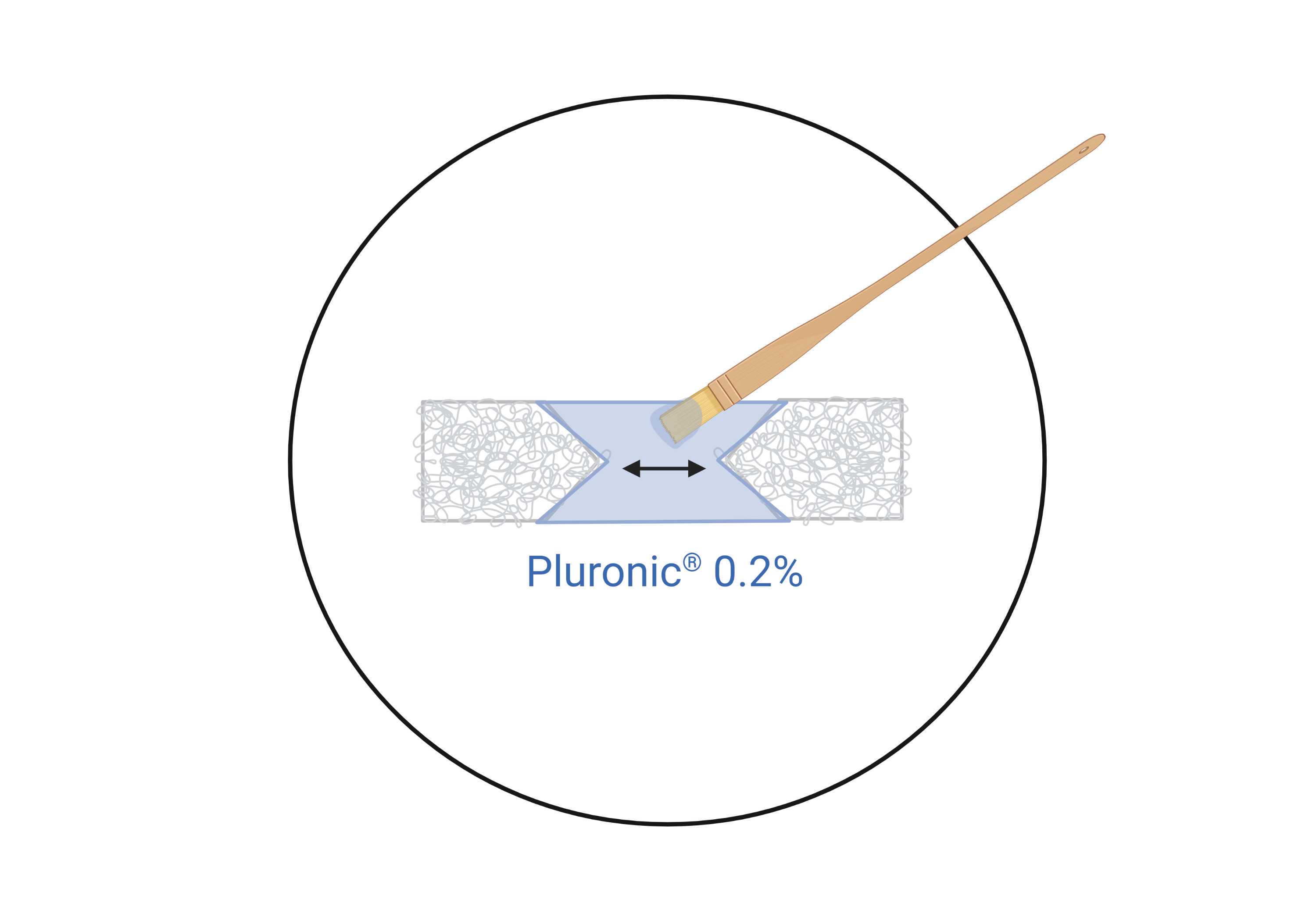
After the plates have been dried in the incubator, perform coating with a sterile solution of 0.2Mass / % volume
In order to coat only the middle region where the muscle bundle will be placed, and avoid soaking the velcro with a hydrophobic material, the Pluronic® solution is applied with a sterile fine paintbrush. Sterilize the paintbrush by immersion in 70% ethanol for 0h 15m 0s, and dry by aspiration with vacuum. Ideally, choose a paintbrush with short and square bristles.
Once all plates are coated, leave the dishes in the incubator (37°C and 5% (v/v) CO2) for 1h 0m 0s . Importantly, during this time, the coating solution will completely evaporate, leaving only the solute on the surface. After this, it is not necessary to aspirate or wash the coating solution; the hydrogel is directly seeded onto the plates. In this way, the plates will remain completely dry at the time of seeding, which will enable correct spreading of the hydrogel as shown in Step 14.5 .
Seeding and differentiation of muscle bundles
Trypsinize cells for seeding according to standard procedures. A large volume of cell suspension may be obtained depending on the number of devices to be seeded, so for comfort, it can be collected in one of the used flasks before counting. From there, take an aliquot of the cell suspension and count the cells.
Then, calculate the volume needed for obtaining 3·106 cells 6 cells, and place this volume into individual Falcon tubes. Each Falcon tube will contain enough cells for one muscle bundle. Centrifuge cells at 800 rpm for 0h 5m 0s .
After centrifuging cells, place the Falcon tubes in the hood and use them sequentially to make muscle bundles. The cells will be resuspended in a hydrogel containing the components listed in Figure 6 . All components must be kept on ice before and during the seeding procedure.
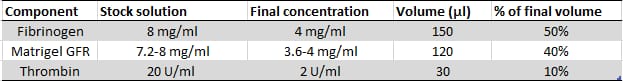
First, aspirate the supernatant and resuspend the cell pellet in the indicated amount of fibrinogen (150µL). Keep the Falcon tube on ice hereafter.
Then, add the indicated amount of Matrigel® (120µL) and homogenize the mixture gently by pipetting.
After this step, prepare a P1000 pipette with a pipette tip and set it to 300µL. Set it aside until needed for seeding.
Add 30µL of thrombin solution to the tube, and then swiftly homogenize the contents of the tube using the P1000 pipette (4-5 times, gently). Aspirate all of the volume and take it to one of the Petri dishes.
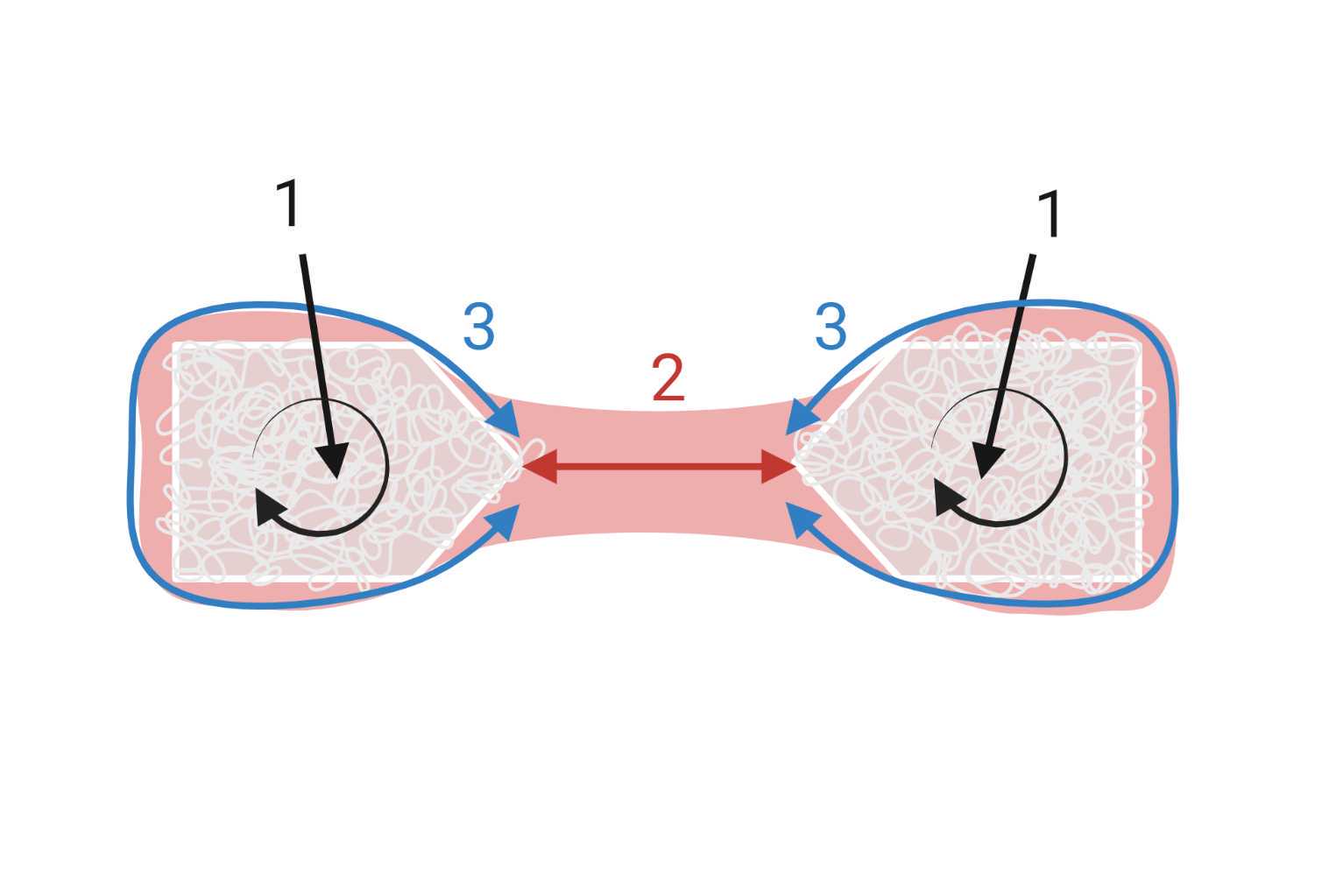
For seeding the hydrogel-cell mixture , quickly but carefully follow the directions in Figure 7 . It is important to follow this order for distributing the liquid onto the VELCRO® and forming the correct shape.
- First, add part of the solution to both VELCRO® anchors, and make sure to push the liquid into the loops by repeatedly pressing with the pipette tip in all directions. Due to the viscosity of the solution, it will not enter into the loops unless pressed into them, and this is important for forming a continuous structure with the middle portion.
- Then, add most of the remaining solution to the middle part, and connect the two VELCRO® anchors to each other.
- Finally, distribute the hydrogel around the anchors to ensure that all sides are covered.
After seeding, the resulting structure will look flat and wide ( Figure 8 ), as the hydrogel is still attached to the plate. Nevertheless, the shape will improve upon differentiation and compaction of the muscle bundles.

After two days in maintenance medium, switch the cells to differentiation medium . In Figure 9 , the composition of differentiation medium for AB1079 cells is shown. This medium is prepared using a 3:1 mixture of DMEM and M199, respectively. Agrin is used for mimicking neural signaling for AChR clustering. 6-Aminocaproic acid (6-ACA) is used as an antifibrinolytic agent, which aids in the long-term maintenance of the structure.
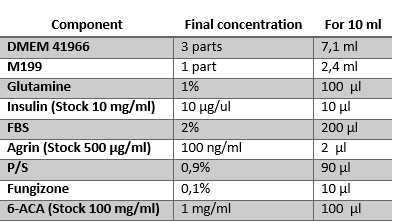
References of used materials:
- DMEM 41966:
- M199:
- Glutamine:
- Insulin:
- FBS:
- Agrin:
- P/S:
- Fungizone:
- 6-ACA:
Change half of the medium every 2 days (or right before and after a weekend), over the course of 14 days. 336h 0m 0s .
Validation of muscle differentiation
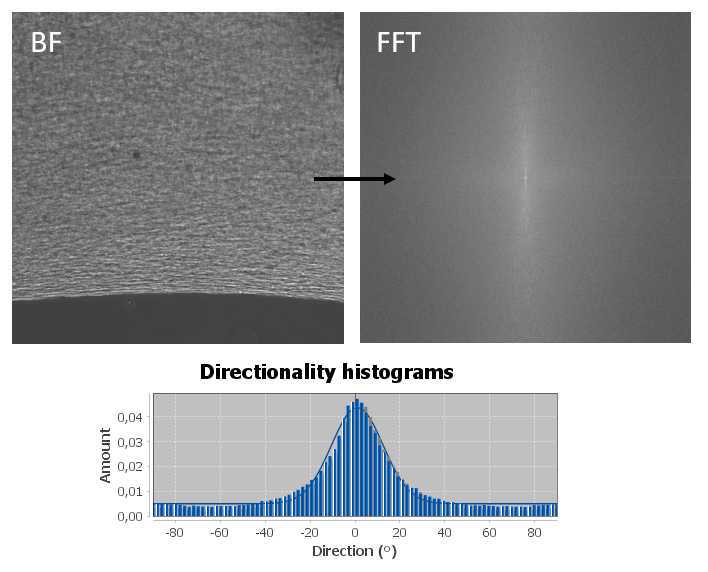
During differentiation, take brightfield images of the muscle bundles at 4X and 10X magnification. With these images, perform a directionality analysis to show alignment of cells. This can be done in ImageJ with the Directionality plugin, which will provide a Fast Fourier Transform (FFT) of the image, and a directionality histogram ( Figure 11 ).
In order to confirm that differentiation has taken place, perform immunostaining of two key markers: Myosin Heavy Chain (MHC) and acetylcholine receptors (AChR). The latter are relevant if it is desired to perform chemical stimulation with acetylcholine. After staining, perform confocal imaging of MHC and AChR. It is recommended to view both of these markers at 40X magnification.
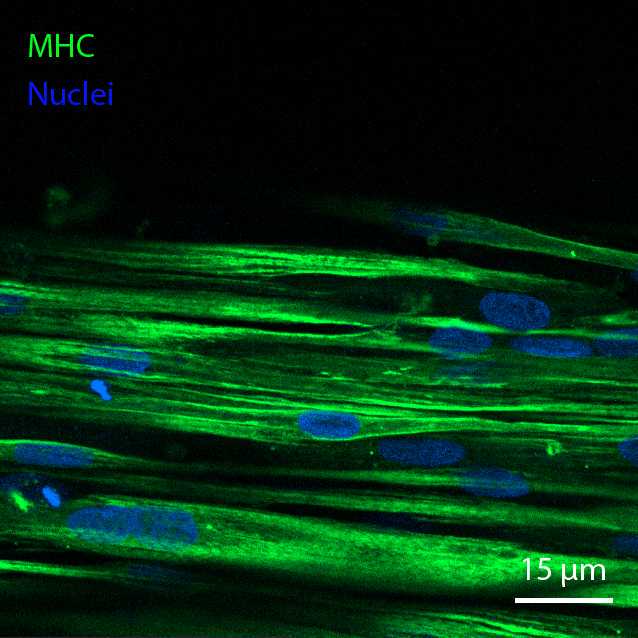
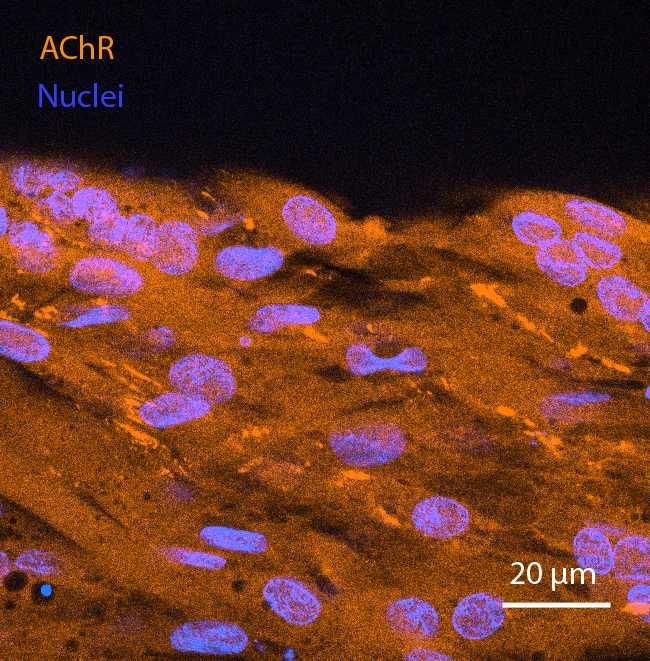
Fix the cells with phosphate-buffered paraformaldehyde (PFA) 4Mass / % volume at Room temperature in a fume hood. First, add one volume of 4% PFA directly to the cell culture medium, to obtain a final concentration of 2% PFA. Incubate for 0h 10m 0s , aspirate, and fix for 0h 30m 0s to 1h 0m 0s in 4% PFA. After fixation, wash thoroughly with PBS (3 times) before taking the plates out of the fume hood.
Block the putative non-specific binding of the antibodies for 6h 0m 0s at Room temperature , in a blocking buffer of PBS - Gelatin 1Mass / % volume with 0.5% (v/v) Triton X-100 and 10% (v/v)
In order to reduce the necessary amount of antibody solutions and enable mounting, cut the muscle constructs using a scalpel as shown in Figure 14. Pick up the cut-out middle portions using a fine paintbrush, and place them in the well of a 24-well plate. Note that the plate and well must contain PBS, in order to facilitate detachment of the hydrogel from the paintbrush.
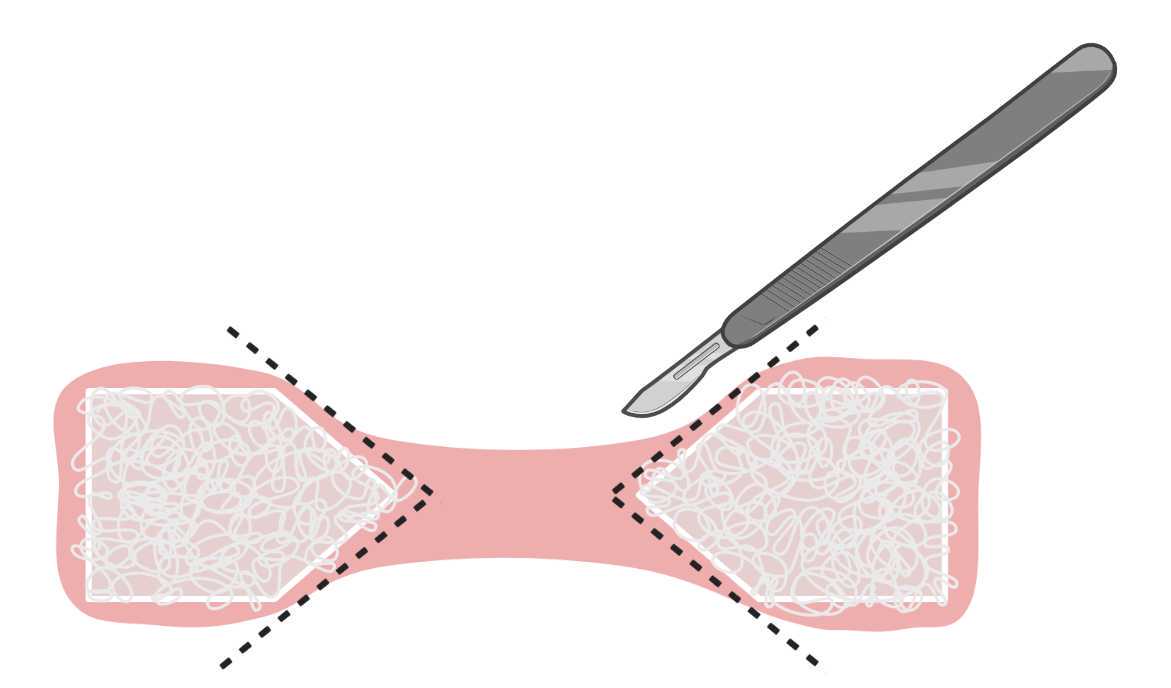
Perform antibody staining. Prepare antibody or staining solutions in PBS - Gelatin 1Mass / % volume with 0.5% (v/v) Triton X-100 and 5% (v/v)
All incubations and washes are performed with gentle shaking. Perform washes (3-4 washes of 0h 10m 0s) between stainings, with PBS containing 0.5% (v/v) Triton X-100.
Antibodies:
- MHC :
1:400 dilution. Incubate for 3 at 4°C, and follow up with, incubated for 6h 0m 0satRoom temperatureto at4°C. - AChR :
1:500 dilution. Incubate for 6h 0m 0satRoom temperatureto at4°C.
Finally, wash the samples 3 times for 0h 10m 0s with PBS containing 0.5% (v/v)Triton-X100, and finally 3 times with PBS.
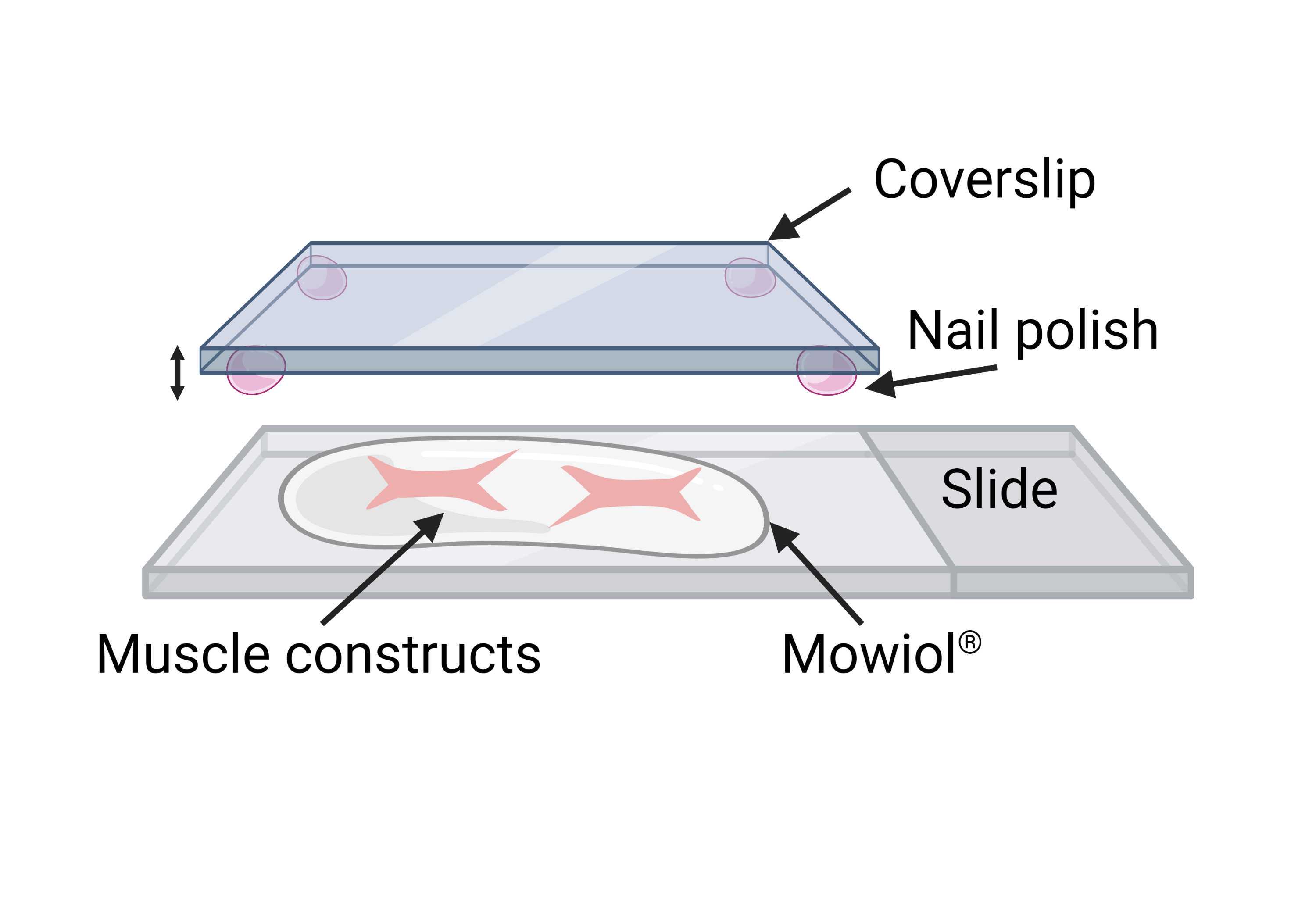
In order to mount the samples for confocal imaging, it is necessary to use coverslips with small drops of nail polish at the corners (prepare in advance and let them dry). The height of the dried nail polish will create a gap for the muscle bundle to be mounted without risk of excessive deformation ( Figure 15 ).
Transfer the muscle bundles onto a glass slide using a fine paintbrush, and cover them with 4°C .
Electrical pulse stimulation
After 14 days in differentiation medium, cells can be stimulated electrically to prove that they have the ability to contract synchronically in response to electrical pulses.
Contractions may be imaged in brightfield and processed using the MUSCLEMOTION plugin for ImageJ, or alternatively, calcium imaging can be performed using Fluo-4 AM and subsequent ΔF/F0 0 analysis.
Before stimulation, it is necessary to build a simple stimulation device , which consists of two graphite electrodes attached to the lid of a 35 mm ∅ Petri dish ( Figure 16 ). The graphite rods are bound to two copper wires, which go through two small holes on the lid of the Petri dish. The holes and wires are sealed together with Loctite® Super glue.
The electrical stimulation setup consists of this stimulation device, an electrical pulse generator (we use Aim TTi TG2512A), and an oscilloscope. The pulse generator is connected to a T-adapter, which connects to the oscilloscope and to two positive and negative cables with alligator clips. The alligator clips are clipped to the wires on the electrodes.
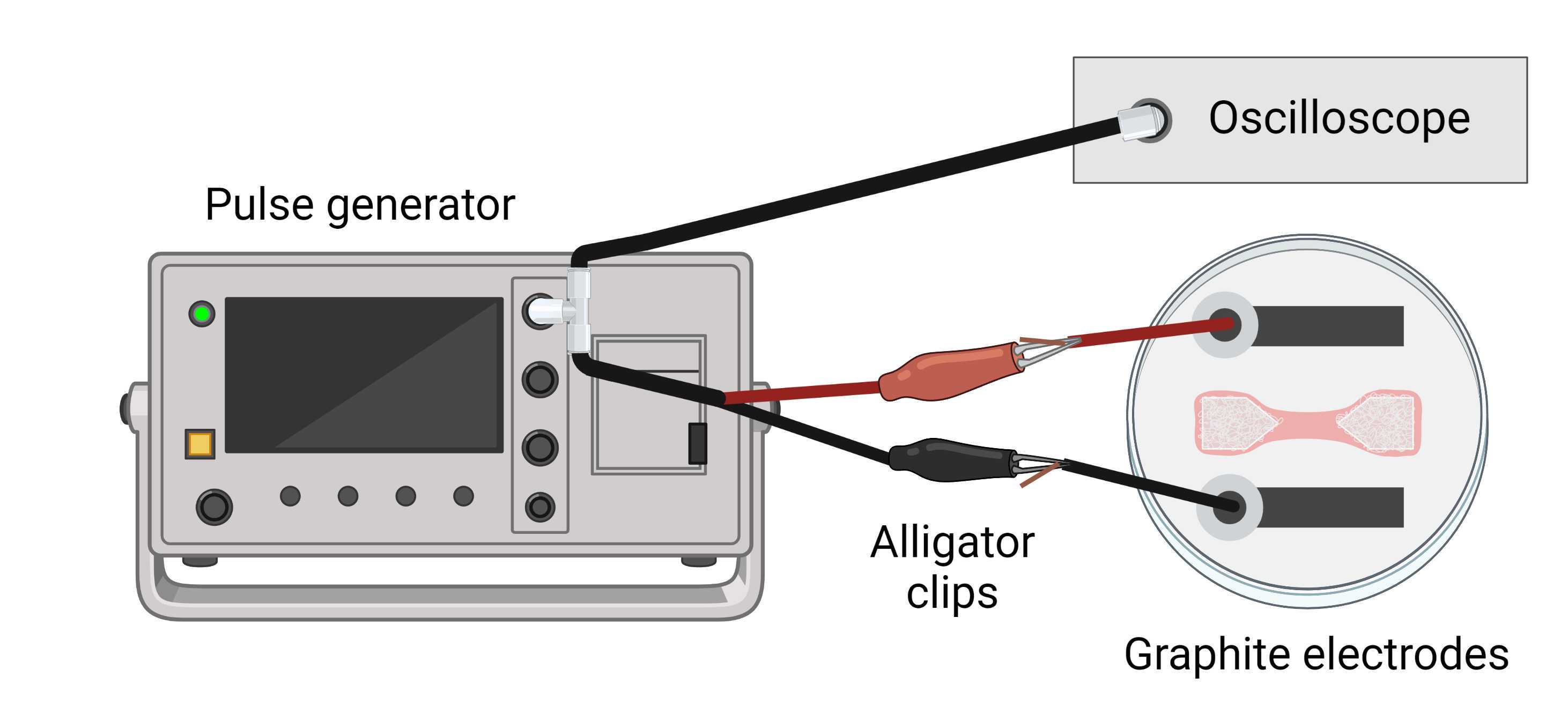
On the day that stimulation is to be performed, if needed to maintain sterile conditions, sterilize the stimulation device by placing the lid onto a 35 mm Petri dish filled with 70% ethanol. Incubate for 0h 15m 0s, wash 3 times with sterile H2O, and then sterilize the lid under UV light for 0h 15m 0s with the electrode part facing upwards.
Inside the biological safety hood, place the electrode lid onto the Petri dish of interest, with the electrodes parallel to the muscle bundle ( Figure 16 ). Take the closed plate to the microscope, and fasten it to the microscope stage using appropriate holder, or alternatively, two pieces of tape placed at the sides. This will avoid accidental opening of the lid when attaching the electrodes to the pulse generator.
For stimulation, the electrodes on the lid are connected to a pulse generator, programmed with the following recommended parameters :
- Pulse duration: 10 ms
- Frequency: 1 Hz
- Amplitude: 10 V (monophasic)
Higher frequencies may be tested if it is desired to study the tetanization of the muscle fiber. Different voltages (5-20V) may be required depending on the cell line in order to see a robust contraction response. Pulse durations between 2-10 ms may also be tested.
The pulse generator is connected to an oscilloscope to verify that the pulses are correctly generated.
Program the microscope settings. Depending on the type of analysis that has been chosen (contraction analysis or calcium transient analysis), the microscope settings will be programmed differently. Go to Step 33 or Step 41 , respectively.
Attach the graphite electrode cables to the pulse generator using alligator clips, and fasten these cables to the microscope stage using a piece of tape, to avoid unwanted movement of the cables.
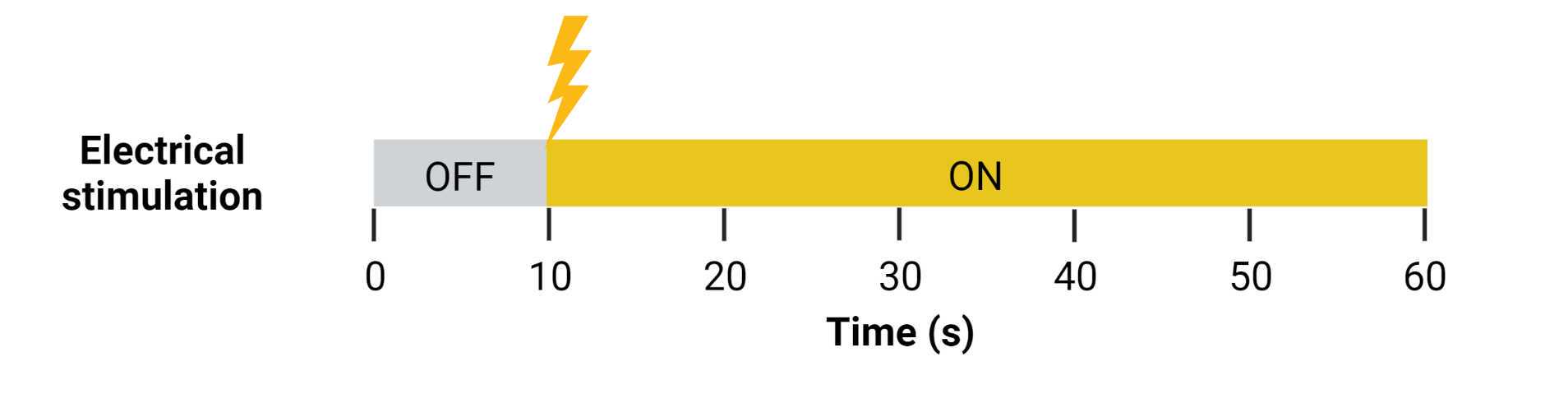
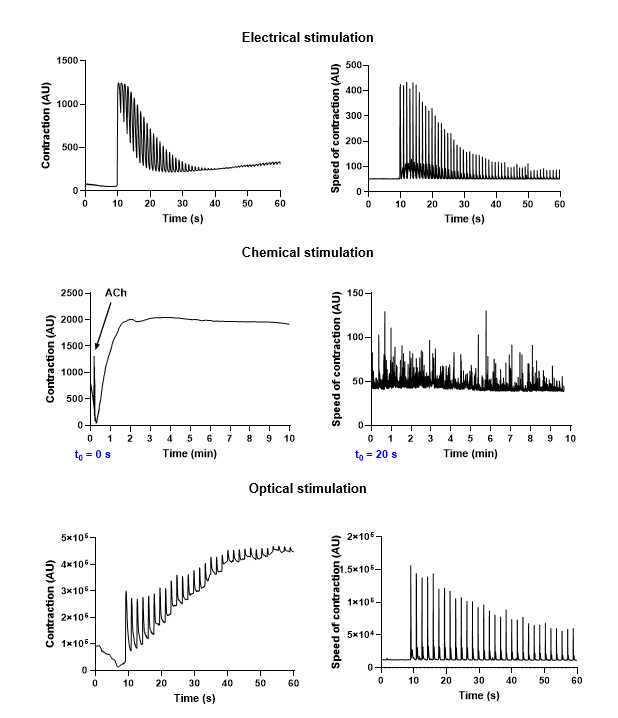
When all the setup is complete, start the video recording . Apply the stimulation regimen shown in Figure 18 : After 0h 0m 10swith no stimulation, turn on the pulse generator and stimulate the muscle bundle for the remaining time of recording (0h 0m 50s). Then, turn off the pulse generator, unfasten the alligator clips, and return the cells to the incubator. Repeat the process with any remaining plates.
Preparation of solutions for chemical stimulation
As a positive control of chemical stimulation, prepare a solution of KCl in Milli-QTM H2O. 1Molarity (M) KCl in Milli-QTM H2O. It is not necessary to keep this solution sterile, although it may be filtered if desired.
On the day of stimulation, prepare a stock solution of acetylcholine in phenol red-free DMEM. Weigh the appropriate amount of acetylcholine chloride to prepare a 100X solution (10millimolar (mM)). For example, for 10mLof stock solution, weigh 18.2mg of 10mL of phenol red-free DMEM
Chemical stimulation with acetylcholine and KCl
Before stimulation with chemical solutions, ensure that the volume inside the Petri dish is 3 ml . If it is lower, change/add medium before stimulation. Medium may have experienced evaporation in the incubator, which can affect the final concentration of the added solutions.
Turn on the microscope. Make sure to place both chemical solutions (ACh and KCl) next to the microscope. Bring pipettes and pipette tips in order to add the chemical solutions when needed.
If the plate contains 3 ml of medium, the amounts to be added of each solution are:
- ACh : 1:100 dilution --> 30 μl --> P100 pipette
- KCl : 1:10 dilution --> 300 μl --> P1000 pipette
Program the microscope settings. Depending on the type of analysis that has been chosen (contraction analysis or calcium transient analysis), the microscope settings will be programmed differently. Go to Step 33 or Step 41 , respectively.
Place the plate at the fluorescence microscope, and focus on the muscle bundle at 20X magnification. It is recommended to focus on the exterior part of the bundle (as in Figure 17 ) which will be more exposed to the chemical stimulant.
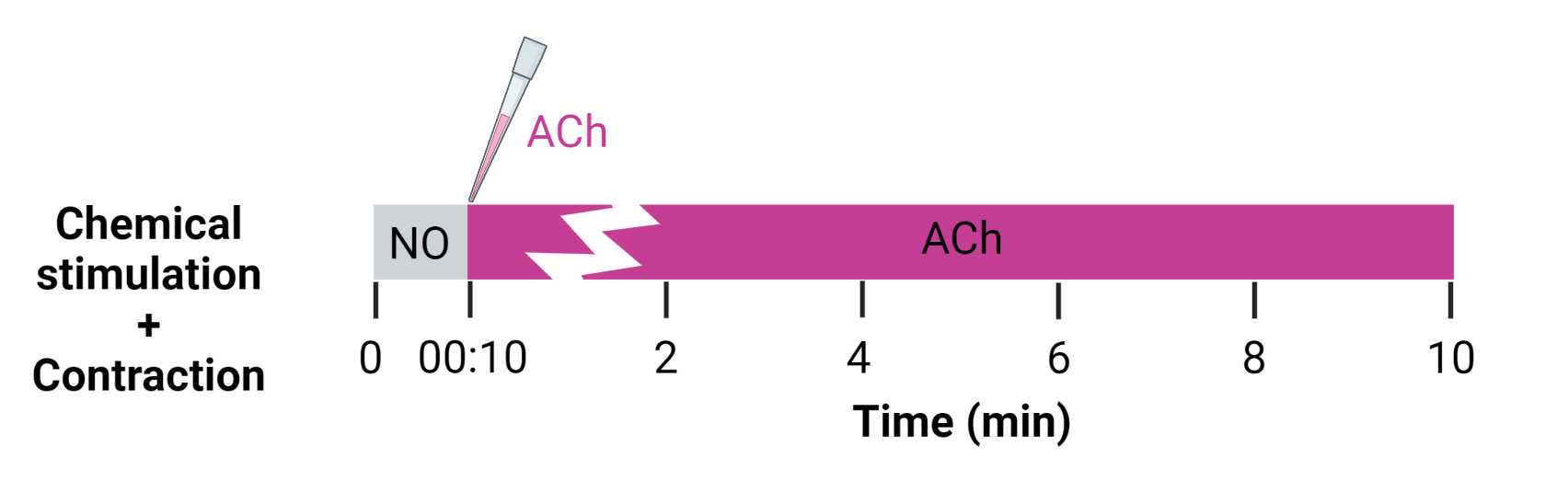
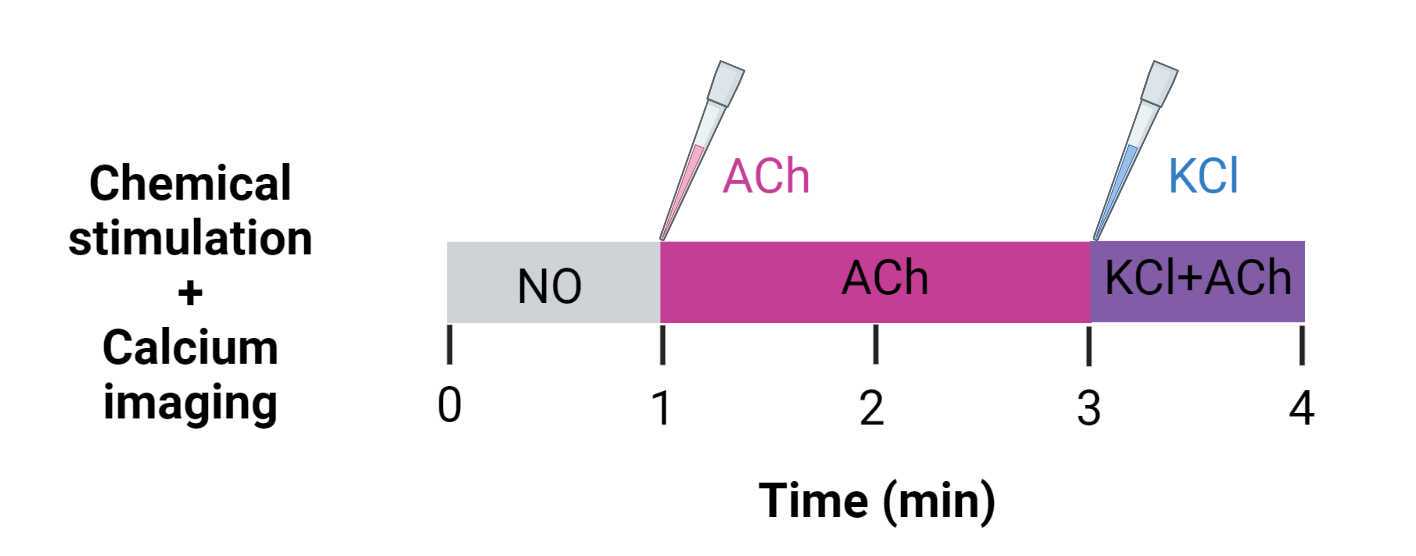
Start the video recording and perform the stimulation according to the chosen stimulation regimen. In our experience, the stimulation regimens shown in Figure 19 and Figure 20 have been appropriate for quantifying the response to the applied stimulation, but may be modified for different needs.
At the chosen times, add the chemical solutions to the medium next to the recorded region, making sure to avoid disturbing the image. For this, pipette quickly (accurate start time of stimulation) but steadily (minimize vibrations in the medium and muscle construct).
Optical stimulation of ChR2+ cells
For optical stimulation, it is necessary to use a myoblast cell line which has previously been made to express channelrhodopsin-2 (ChR2). We have generated ChR2+ human myoblasts by viral infection and FACS purification, as described in a previous publication from our group:
After 14 days in differentiation medium, cells expressing ChR2 can be optically stimulated. Contractions are imaged in brightfield and processed using the MUSCLEMOTION plugin for ImageJ.
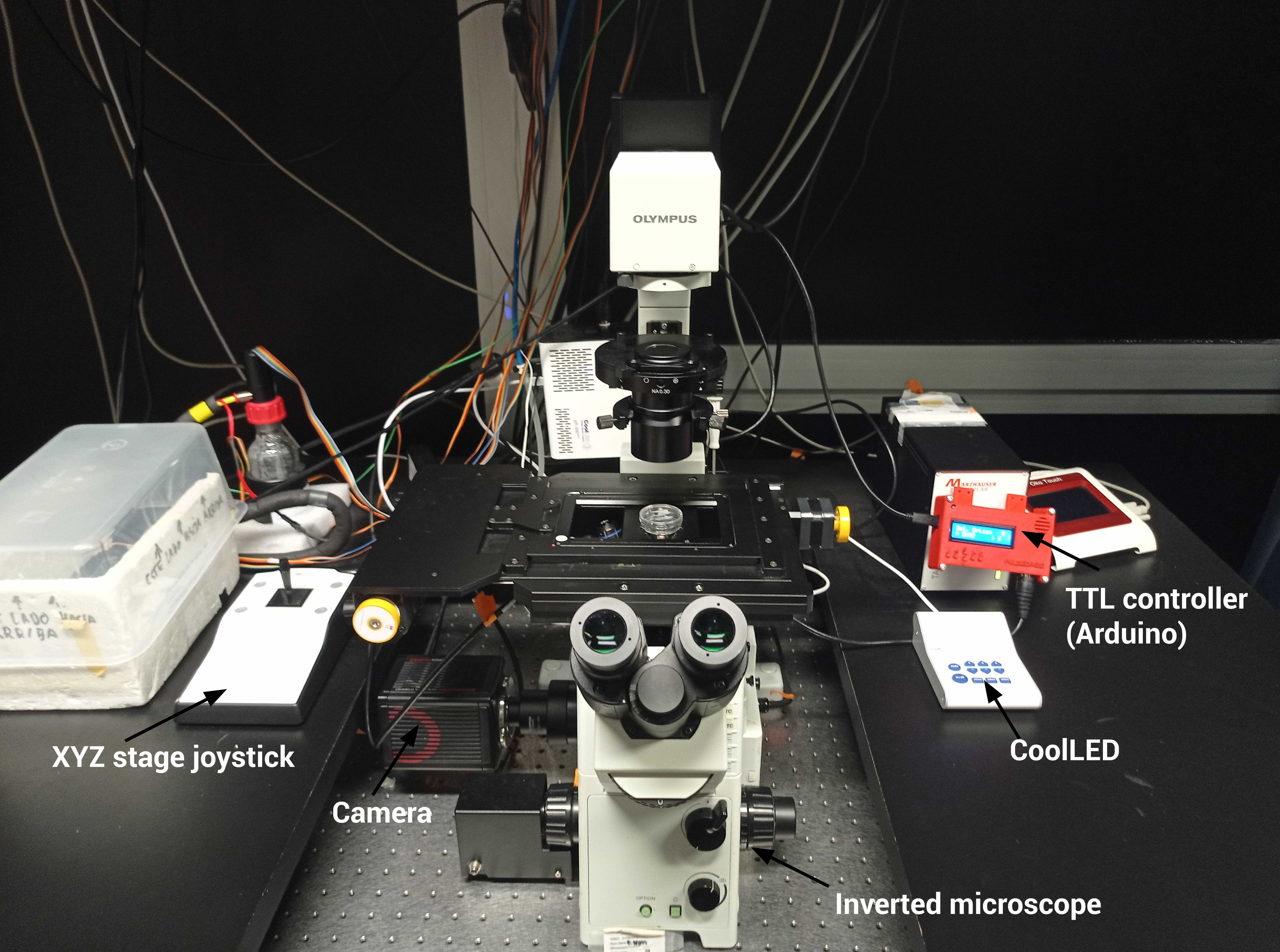
The optical stimulation setup consists of a coolLED pE-300 illumination system coupled to an inverted microscope (i.e., Olympus IX71) to visualize the muscle contraction. The TTL control of the illumination system allows for the delivery of precisely timed sequences of light. The TTL can be directed to an Arduino-UNO™ microcontroller pulse generator or alternatively a PulserPlus generator and Pulser v3.1 software (Prizmatix, Israel). The average light intensity with this setup is ≈ 20-25 W/cm2 , measured at the culture dish with a Newport 1919 optical power meter (Newport Photonics, USA).
The two alternative setups are shown in Figure 21 and Figure 22 , respectively.
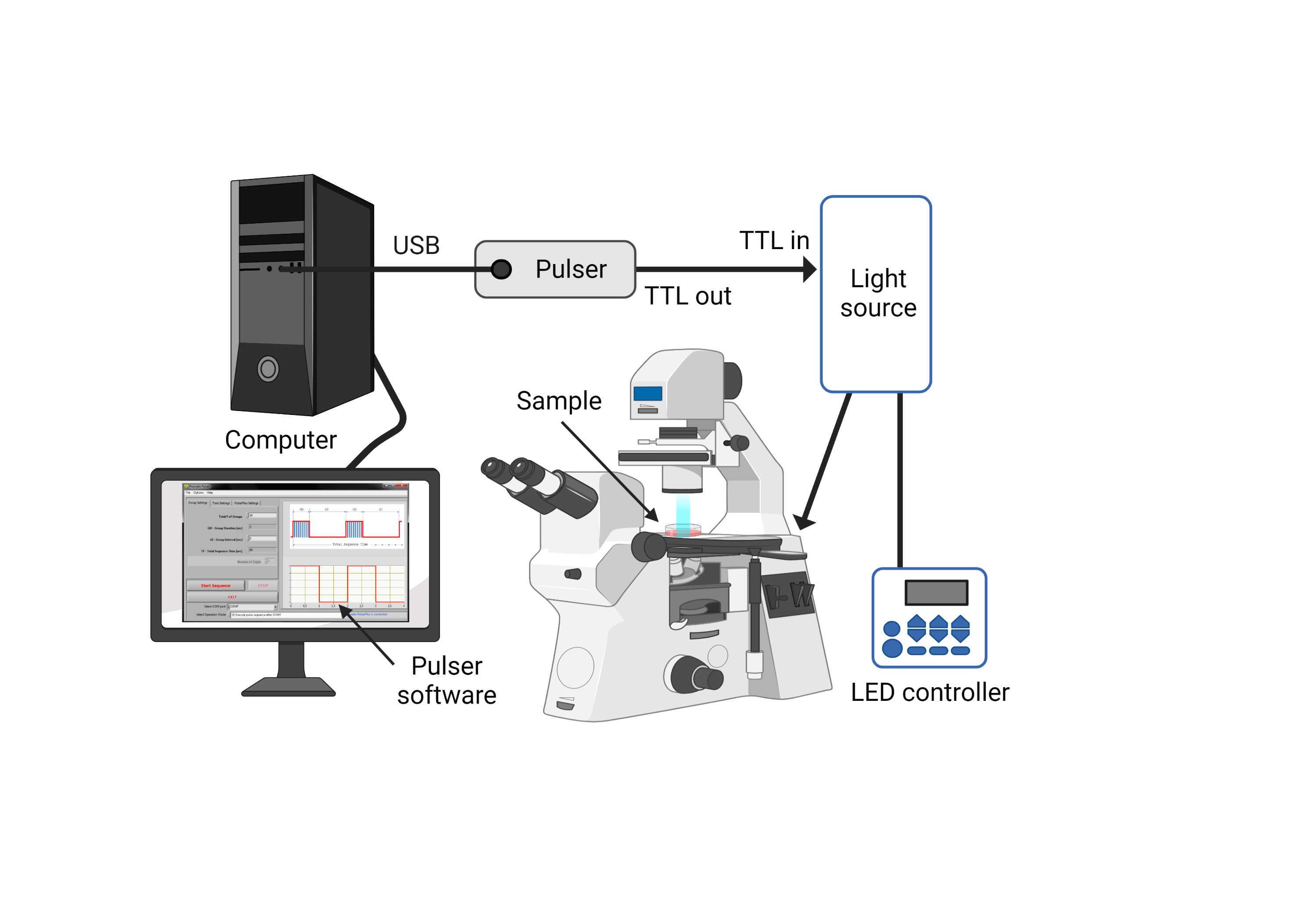
For optical stimulation, turn on the Arduino or the pulse generator, and program the following recommended parameters :
- Pulse duration: 5 ms
- Frequency: 20 Hz
- Train duration: 1 s ON and 1 s OFF. (During the 1s when the stimulation is ON, 20 pulses/s are applied).
- Total duration: 1 min (30 repetitions of 1s ON + 1s OFF).
Program the microscope software to record a brightfield video with the parameters described in Step 33 .
Turn on the bright field microscope illumination. Set up the image capture in live view. Fine-tune imaging positions and focus using the microscope software before starting the experiment. Use a microscope objective which allows the observation of individual cells (20X in our experiments).
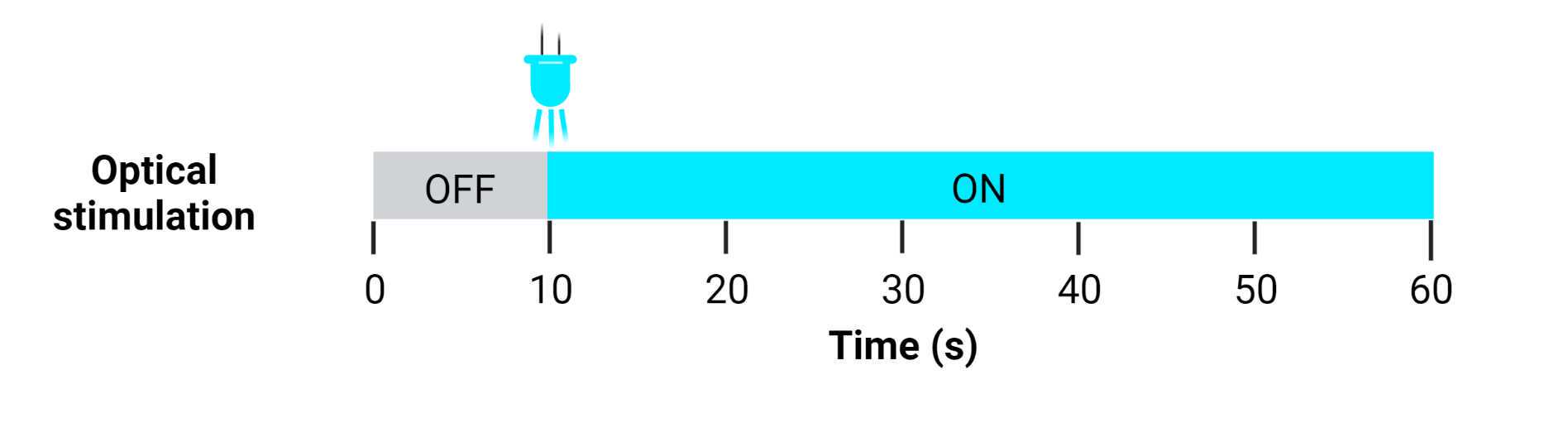
When all the setup is complete, start the video recording. Follow the stimulation regimen shown in Figure 23 : After 0h 0m 10s with no stimulation, turn on either the Arduino or the pulse generator and stimulate the muscle bundle for the remaining time of recording (0h 0m 50s). Then, turn off the pulse generator and return the cells to the incubator. Repeat the process with any remaining plates.
Contraction analysis with MUSCLEMOTION
If performing contraction analysis, program the following **recommended microscope settings for brightfield videos** :
- Resolution: 1024x1024 px, 16-bit
- Total time of recording: 1 min or more, depending on chosen stimulation regimen
- Frame speed: 40 fps (1 image every 25 ms)
- Exposure time: lower than 25 ms, check that the image acquisition time is faster than the desired frame speed.
Regarding software, commercial (such as Olympus cellSens) or open-source software (μManager) can be used to program these parameters.
After brightfield imaging of contraction, analyze the contraction response with the MUSCLEMOTION plugin for ImageJ . This protocol can be used for electrical, chemical or optical stimulation videos.
If the videos are obtained in certain microscope software formats such as .vsi, it will be necessary to open them with the Bioformats plugin (Bioformats Importer), and then save them as a .tif "Image Sequence". Opening and converting large videos may be time-consuming depending on your computer's processing speed.
Open the MuscleMotion plugin in ImageJ and click "Run". Select the frame rate according to the frame rate in the input video.
Leave all other parameters as shown in Figure 24 and click OK. This will open a file selection menu to select the output directory (recommended to create a new folder) and the input directory (the folder with the TIFF image sequence).

After running the program, the processing may take several minutes to be completed. The files that will be obtained in the output folder are:
- Reference frame
- Contraction: graph and text file
- Contraction speed: graph and text file
The text files contain contraction or contraction speed information at each frame, which can be used for creating a graph in any other software of your choice (for example, GraphPad Prism, as in Figure 25 ).
Calcium imaging: Fluo-4 AM loading and deesterification
Calcium imaging may be combined with electrical or chemical stimulation protocols.
Fluo-4 AM sterile stock solution should be prepared in advance at 1millimolar (mM) in DMSO with 20Mass / % volume Pluronic®, and stored at -20ºC protected from light.
To prepare the solvent, dissolve 2g of Pluronic® in 10mL of sterile DMSO37°C water bath for 0h 20m 0s min and vortex the solution until there are no more clumps of Pluronic®. Then, filter this mixture at 0.22 μm in the biological safety hood.
To reconstitute Fluo-4 AM, follow the manufacturer's instructions.
Wash cells three times with pre-warmed phenol red-free DMEM (
Add Fluo-4 AM at a final concentration of 10micromolar (µM) in pre-warmed phenol red-free DMEM.
For each Petri dish, use 2mL of medium (enough to cover the muscle bundle) and 20µL of Fluo-4 AM stock solution (1millimolar (mM) in DMSO with 20Mass / % volume Pluronic®). Perform loading of the Fluo-4 AM in the incubator for 1h 0m 0s .
After loading is completed, . Then, for deesterification of Fluo-4 , leave cells in phenol red-free DMEM at roomRoom temperature for 0h 15m 0s and then at 37°C for another 0h 15m 0s .
Proceed to imaging shortly after deesterification . Fluorescence intensity and cell viability decrease after prolonged times in the incubator. Therefore, if imaging many samples on the same day, it is necessary to perform loading and imaging in a staggered manner.
Recommended fluorescence microscope settings :
- Total video time: depending on the type and regimen of stimulation.
- Magnification: 20X
- Fluorescence: ~70-80s exposure time, blue light (adjust for your microscope/lamp/cells). Must be lower than the acquisition time.
- Acquisition time: 100ms (10 fps).
Calcium transient analysis
In order to analyze videos obtained from Fluo-4 AM calcium imaging experiments, we use ImageJ for processing and obtaining ΔF/F0 data 0 data. In order to analyze calcium transients, individual cells are manually delimited using the ImageJ ROI manager. Then, fluorescence intensity data is extracted at each timepoint. This data is normalized to the mean basal fluorescence.
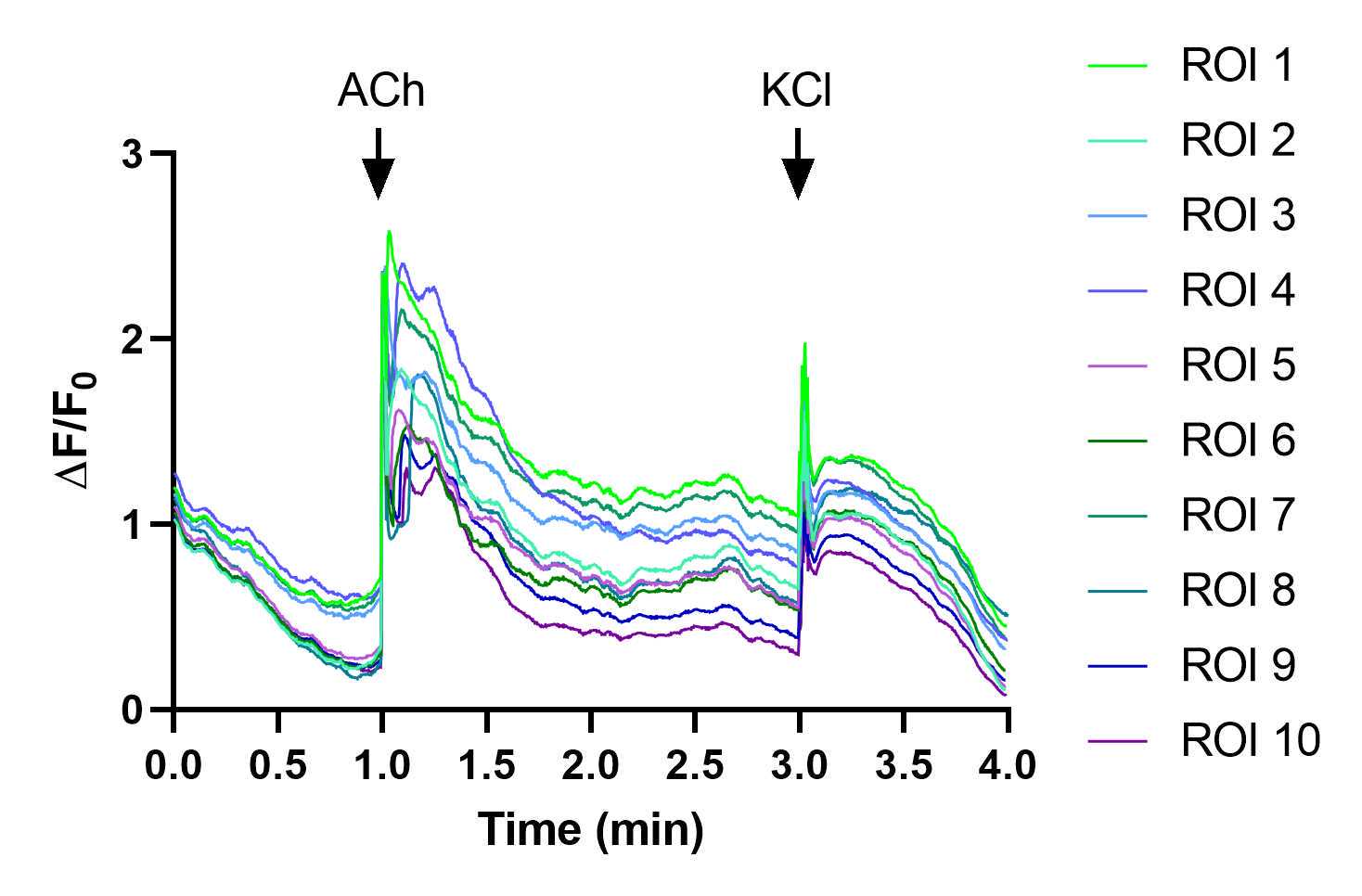
As an example, a video using chemical stimulation with 100micromolar (µM) ACh and 100millimolar (mM) KCl is analyzed in the following steps. This video was recorded at 100 ms/frame (10 fps), for a total of 4 minutes.
First, open the video file in ImageJ . Play the video file using the sliding bar, and identify the timepoint or frame at which the stimulation was initiated.
For example, in the case of a video with the stimulation regimen shown in Figure 20 (with addition of acetylcholine and KCl), key timepoints would be 1 min (60000 ms = 600 frames) for ACh addition and 3 min (180000 ms = 1800 frames) for KCl addition, respectively.
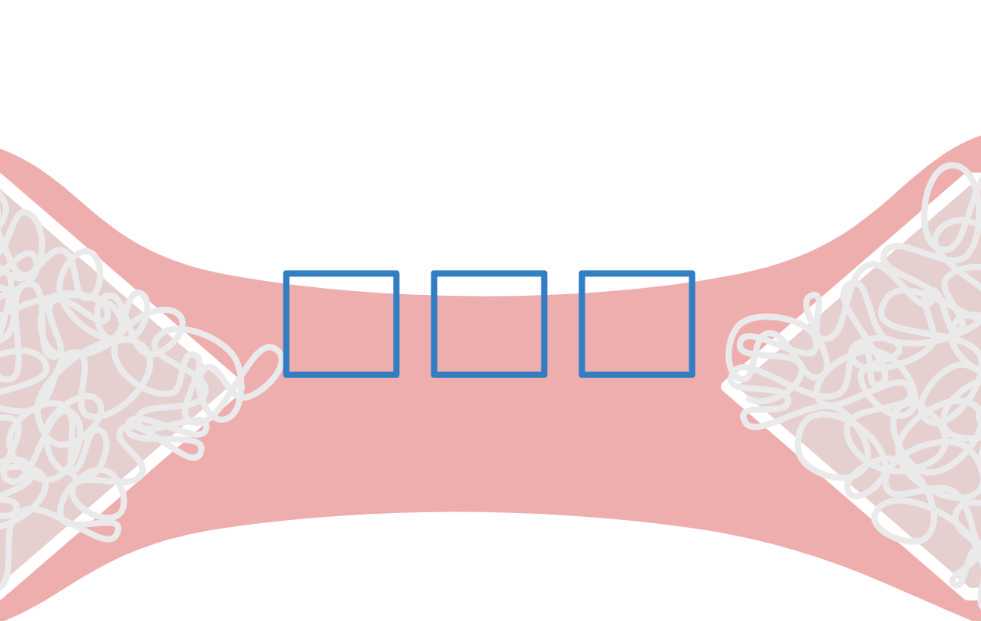
By moving the sliding bar back and forth, visually identify cells that respond to the stimulation after the start of stimulation (see differences between Figure 26 and Figure 27 ). These cells will later be selected as regions of interest (ROIs), as explained in the following steps.
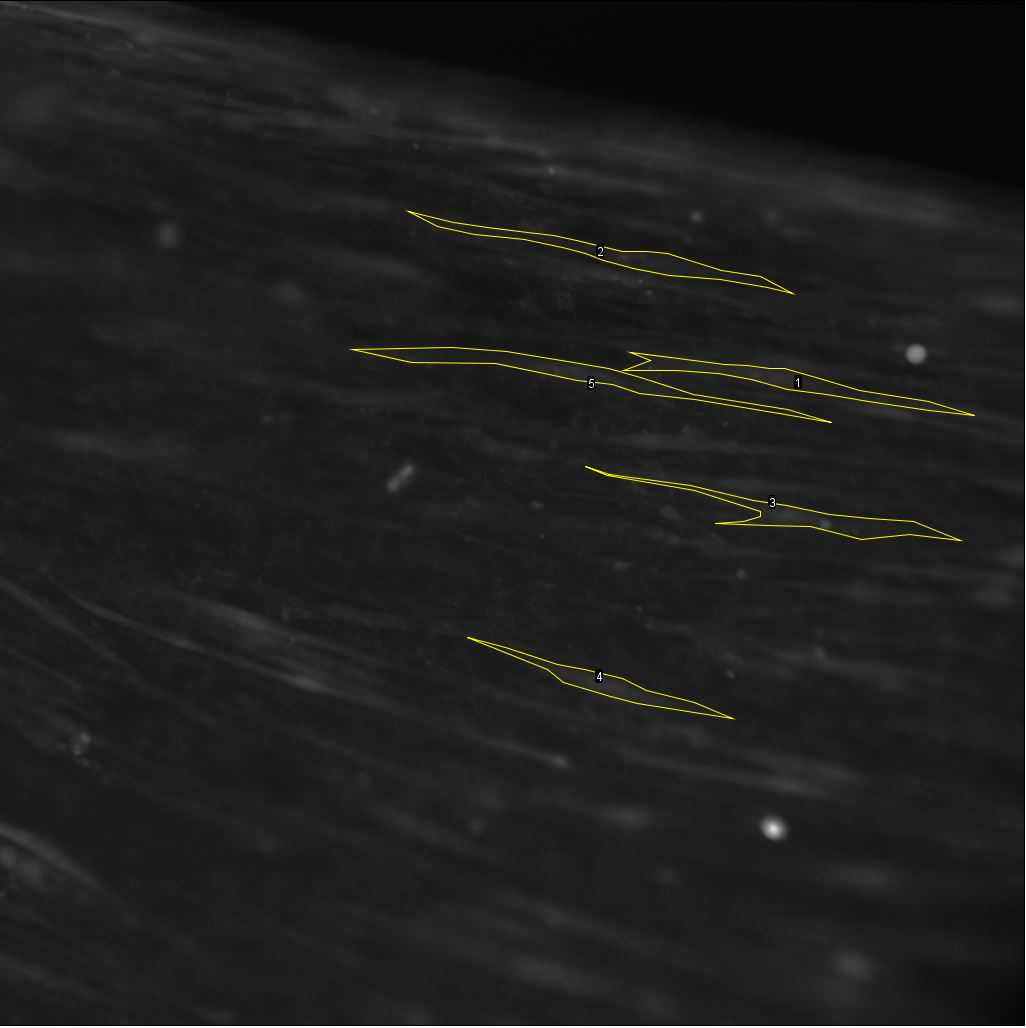
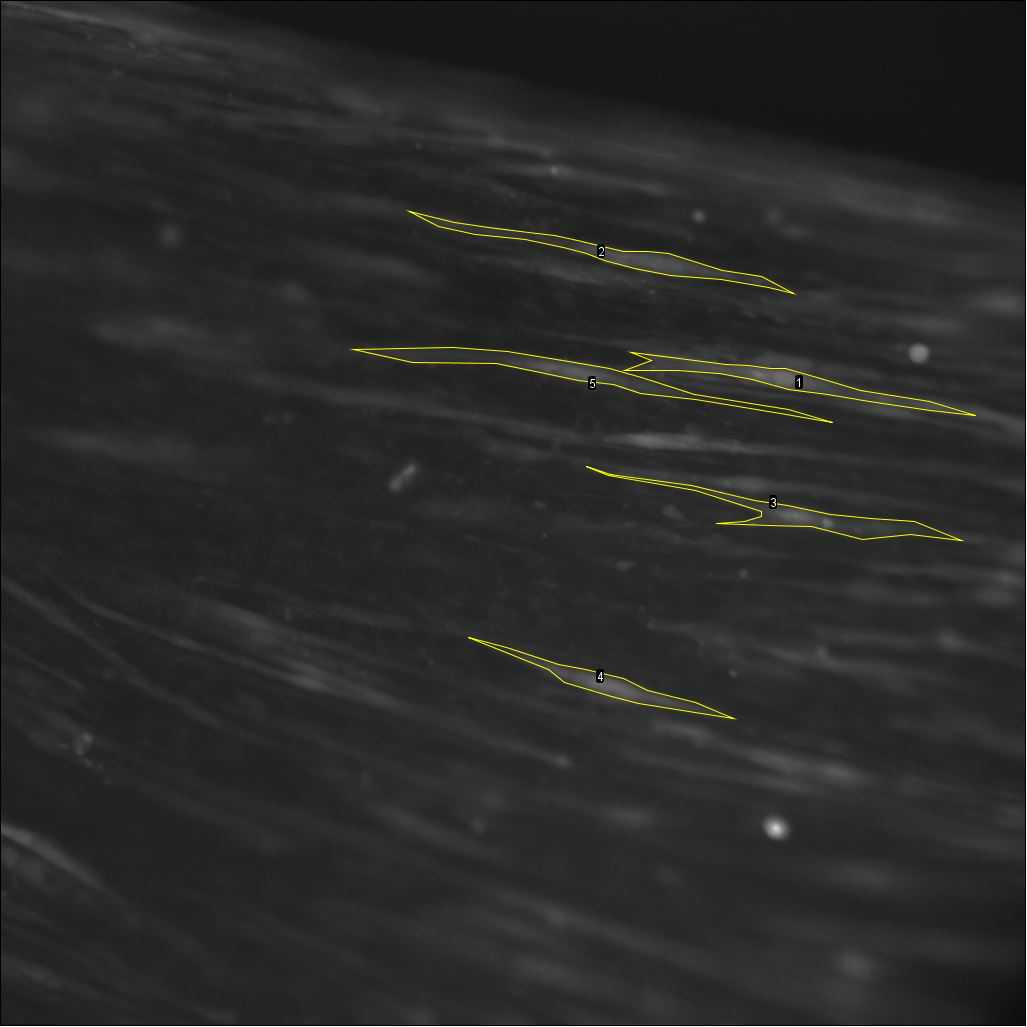
After visually identifying the cells, proceed to define ROIs of a subset of cells (5-10 cells per video) and calculate their fluorescence intensity over time .
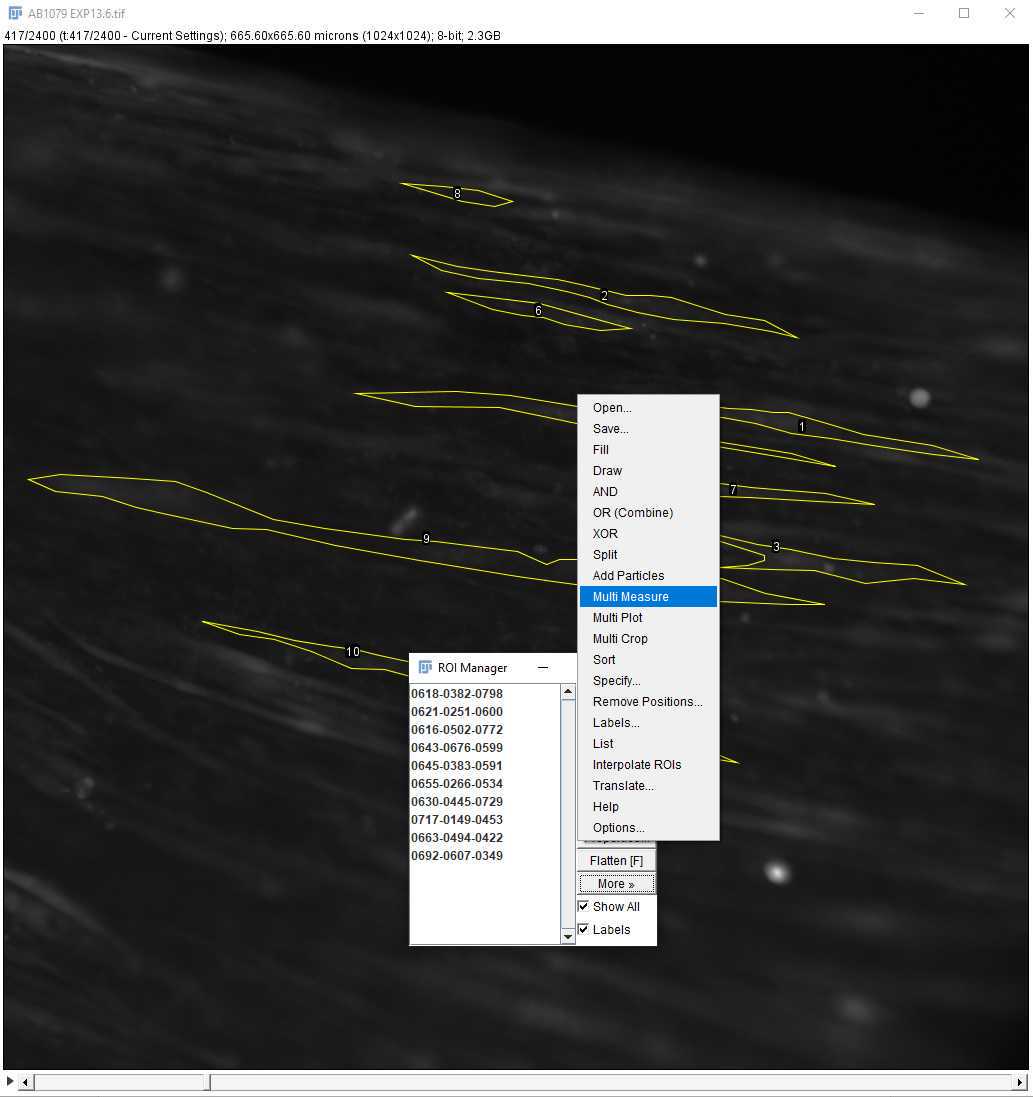
On the ImageJ toolbar, open the ROI manager. Go to Analyze > Tools > ROI manager.
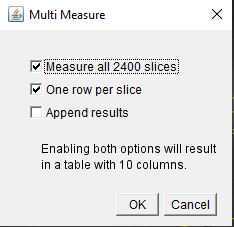
Using the polygon selection tool , outline each of the cells of interest as in Figure . After closing each outline, on the ROI manager, click on "Add" before outlining the next cell. To see all the ROIs, click on "Show all" At the end of the selection, the ROI set can be saved using More >> Save.
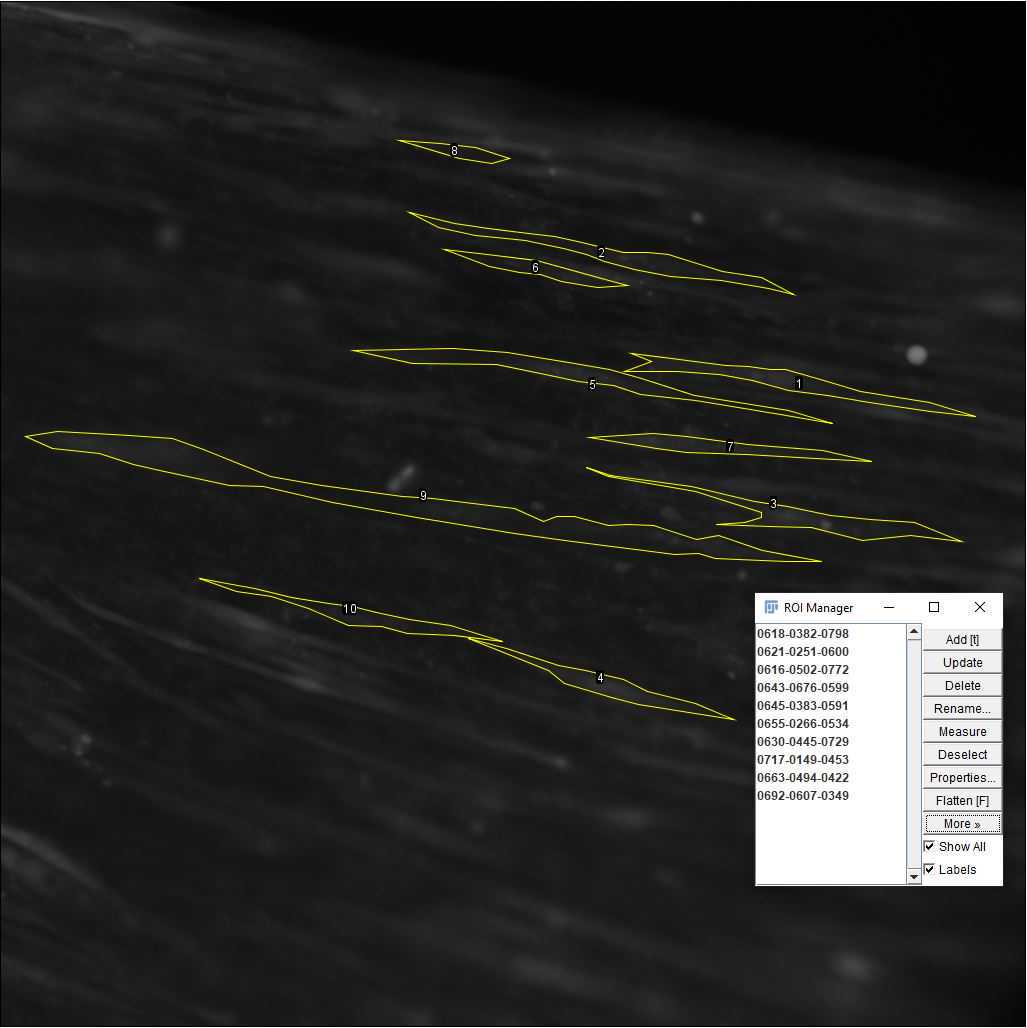
Once the ROIs are selected and saved, go to Analyze > Set Measurements and select "Mean gray value" only ( Figure 29 ). In this way, only the fluorescence information will be extracted from the ROIs. Click OK.
<img src="https://static.yanyin.tech/literature_test/protocol_io_true/protocols.io.b4qiqvue/mk9zbphz732.jpg" alt="Figure 29. "Set measurements" window. " loading="lazy" title="Figure 29. "Set measurements" window. "/>
After the measurements are completed, the results will pop up in a Results window. Each column contains the fluorescence intensity data for one ROI at all the timepoints (each row is one frame). Save these results and/or copy them to an Excel file (or other similar spreadsheet software).
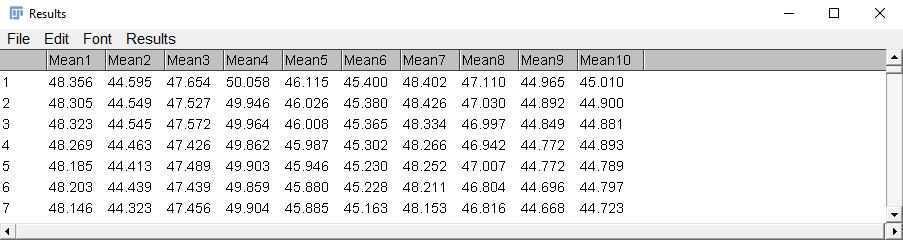
Normalize the fluorescence data to the basal fluorescence (F0) 0) in order to calculate ΔF/F0. 0.
While this ROI is selected, go to Analyze > Measure in order to obtain the fluorescence value for this ROI. Save the result that pops up (this will be F0 0).
Use a software of your choice to generate an XY plot of the ΔF/F0 0over time. In our case, we use GraphPad Prism software, but other open-source software such as Veusz can be used instead.
- X values: time, in increments of 100 ms, or as many ms as separate each frame.
- Y values: ΔF/F0 of each ROI (plot all ROIs in the same graph).
In order to visualize the point at which the stimulation was applied, arrows can be added as annotations. It is also helpful to show each ROI in a different line color.
After these steps, the data generated with this protocol can be used in further analyses that may be interesting for every reserarcher's purposes.