Determine the Size of Sonicated Samples and the DNA Concentration
Vasso Makrantoni, Daniel Robertson, Adele L. Marston
Chromatin immunoprecipitation
Saccharomyces cerevisiae
Schizosaccharomyces pombe
Cohesin
Condensin
Mitosis
Meiosis
Scc1
Rec8
Brn1
Abstract
A plethora of biological processes like gene transcription, DNA replication, DNA recombination, and chromosome segregation are mediated through protein–DNA interactions. A powerful method for investigating proteins within a native chromatin environment in the cell is chromatin immunoprecipitation (ChIP). Combined with the recent technological advancement in next generation sequencing, the ChIP assay can map the exact binding sites of a protein of interest across the entire genome. Here we describe a-step-by step protocol for ChIP followed by library preparation for ChIP-seq from yeast cells.
Before start
Use sonicated chromatin samples (step 15 from "Cell Lysis and Sonication") to determine the fragment size.
Steps
Determine the Size of Sonicated Samples and the DNA Concentration
To a 100µL add 80µL containing 300millimolar (mM).
Decross-link at 65°C 0h 0m 30s.
Add 2µL and incubate at 37°C for 1h 0m 0s.
Add 20µL and incubate at 65°C for 2h 0m 0s.
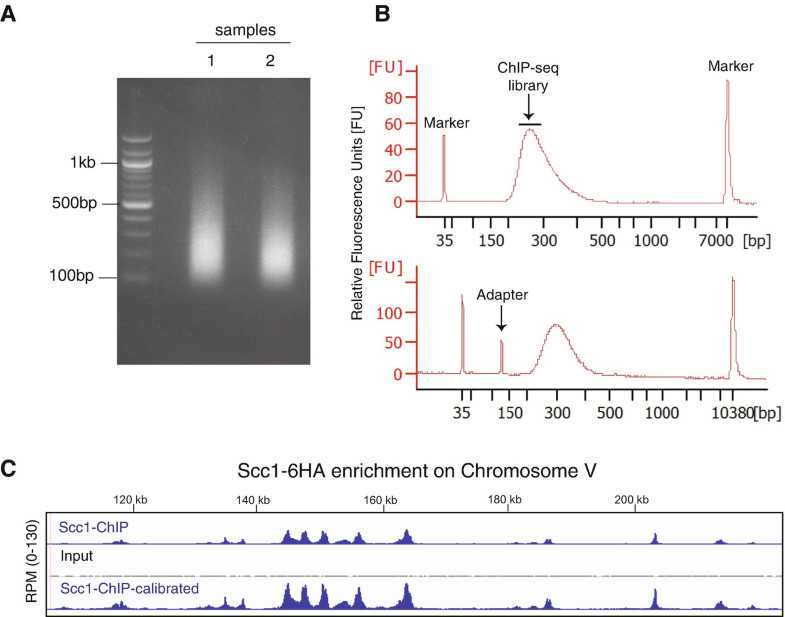
Purify DNA using a PCR purification kit. Run purified DNA on a 2% agarose gel with a 100 bp DNA ladder marker to determine fragment size. Ideally sonication should yield an enrichment of fragments between 200 and 400 bp (Fig. 2a).
Measure DNA concentration using a Qubit HS assay kit.

