Construction of individual ddRAD libraries
Claire Daguin Thiebaut, Stephanie Ruault, Charlotte Roby, Thomas Broquet, Frédérique Viard, Alan Brelsford
Abstract
This protocol describes a double digested restriction-site associated DNA (ddRADseq) procedure, that is a variation on the original RAD sequencing method (Davey & Blaxter 2011), which is used for de novo SNP discovery and genotyping.
This protocol differs from the original ddRADseq protocol (Peterson et al 2012), in which the samples are pooled just after the ligation to adaptors (i.e. before size selection and PCR).
The present ddRAD protocol as been slightly adapted from Alan Brelsford's protocol published in the supplementary material of this paper:
Brelsford, A., Dufresnes, C. & Perrin, N. 2016. High-density sex-specific linkage maps of a European tree frog ( Hyla arborea ) identify the sex chromosome without information on offspring sex. Heredity 116, 177–181 (2016). https://doi.org/10.1038/hdy.2015.83
In the present protocol, all samples are treated separately, in a microplate, until final PCR amplification performed before pooling. Despite being slightly more costly and time-consuming in the lab, it allows for fine adjustement of each sample representation in the final library pool, ensuring similar number of sequencing reads per sample in the final dataset.
Briefly, genomic DNA from the samples are individually digested with 2 restriction enzymes (one rare-cutter and one more frequent cutter) then ligated to a barcoded adaptor (among 24 available) at one side, and a single adaptor at the other side, purified with magnetic beads, and PCR-amplified allowing the addition of a Illumina index (among 12 available) for multiplexing a maximum of 288 sample per library. Samples are then pooled in equimolar conditions after visualisation on an agarose gel. Purification and size selection is then performed before final quality control of the library and sequencing.


Steps
Preparation of double-stranded barcoded P1 adaptors 1µM
In a PCR plate wells, combine each oligo 1.1 with its complementary oligo 1.2 :
5µL adaptor P1-1 10µM
5µL adaptor P1-2 10µM
5µL annealing buffer 10X (100 mM Tris-HCl, pH 8, 500 mM NaCl, 10 mM EDTA Na2)
35µL nuclease free water
Seal the plate with a thermoseal film and incubate in a thermocycler or microplate incubator at 95°C for 0h 5m 0s, and then cool at a rate of 0.1°C / s down to 20°C.
Store at 4°C (or at -20°C for a long-term storage)
| A | B | C | D | E | F | G | H | I | J | K | L |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 9 | 17 | |||||||||
| 2 | 10 | 18 | |||||||||
| 3 | 11 | 19 | |||||||||
| 4 | 26 | 20 | |||||||||
| 5 | 13 | 21 | |||||||||
| 6 | 14 | 22 | |||||||||
| 7 | 15 | 23 | |||||||||
| 8 | 16 | 24 |
Example of a plate map for barcoded P1 adaptors. Allow enough space betweeen the rows to avoid cross-contaminations between barcodes.
Preparation of double-stranded P2 Adaptor P2_Mse 10µM
in a microtube, combine oligo 2.1 with its complementary oligo 2.2
80µL P2-1 100µM
80µL P2-2 100µM
80µL annealing buffer 10X (100 mM Tris-HCl, pH 8, 500 mM NaCl, 10 mM EDTA Na2)
560µL nuclease free water and mix by pipetting
Then aliquot this volume into 100µL in each well of a 8- PCR tube strip.
In a thermocyler or microplate incubator, incubate at 95°C for 0h 5m 0s, and then cool at a rate of 0.1°C / s down to 20°C.
Store at 4°C (or -20°C for a long-term storage)
Genomic DNA preparation
In a PCR plate, put around 50-100ng of genomic DNA in a volume of 30µl (in nuclease free water or Tris-HCl 5mM pH 8.5) for each sample. See guidelines for more details. Ideally, DNA should be free from RNA. For this, include a RNAse treatment in the DNA extraction procedure. If possible, randomize the location of samples in the microplate. Keep a few empty wells for negative controls.
Double digestion
Thaw and vortex all reagents, except enzymes (stored at -20°C), for approximately 0h 0m 5s
Spin down all reagents for approximately 0h 0m 5s and place on ice.
In a microtube, prepare the master mix, according to the following table for a total volume of 35μL:
| A | B | C | D | E |
|---|---|---|---|---|
| concentration | final quantity | N=1 | N=104 (1 plate) | |
| genomic DNA | 50-200ng | 30 | ||
| Cutsmart buffer | 10X | 1X | 3.5 | 364 |
| Enzyme 1 Pst HF | 20u/µL | 8U | 0.4 | 41.6 |
| Enzyme 2 MseI | 10u/µl | 2U | 0.2 | 20.8 |
| H2O | 0.9 | 93.6 | ||
| TOTAL | 35 |
Digestion mix composition
Vortex mix the master mix and spin down.
Aliquot 65µL of the master mix in each well of a 8-PCR tube strip.
In the DNA plate (containing 30µL per well), add 5µL of Master mix with a multichannel pipette and mix by pipetting, seal PCR plate and spin down.
Incubate at 37°C 10h 0m 0s
then 65°C 0h 20m 0s in order to inactivate enzymes
4°C
Check digestion on an agarose gel
Adaptor ligation
Vortex mix all reagents, except enzymes (stored at -20°C), for approximately 0h 0m 5s
Spin down all reagents for approximately 0h 0m 5s and place on ice.
Setup the ligation reaction according to the following table (total volume of 40μL):
| A | B |
|---|---|
| N=1 / µl | |
| digest DNA | 30 |
| ds barcoded adaptor P1 1µM | 2.8 |
| ligation mix (see below) | 7.2 |
In a microtube, prepare the ligation mix according to the following table :
| A | B | C | D | E |
|---|---|---|---|---|
| concentration stock | final concentration | vol 1x µl | Vol µl plate | |
| ds nonbarcoded adaptor P2 | 10µM | 1.8 | 187.2 | |
| Cutsmart buffer | 10X | 1X | 1 | 104 |
| ATP | 10mM | 1 mM | 4 | 416 |
| T4 DNA ligase | 400u/µl | 0.4 | 41.6 | |
| TOTAL | 7.2 |
Vortex the master mix and spin down.
Aliquot 93.5µL of the master mix in each well of a 8-PCR tube strip.
In the DNA digestion plate (30µL per well), add 2.8µL of Adapt P1 according to your plate map, from the adaptor plate (see step 1), with a multichannel pipette.
Then in all wells, add 7.2µL of ligation mix and mix by pipetting, seal PCR plate and spin down.
Incubate at 16°C 6h 0m 0s .
then
4°C
or at -20°C if not performing the bead purification the day after.
Bead purification
This protocol can be used to remove contaminants, unligated adapters, enzymes, buffer additives, salts... and short DNA fragments. The method utilizes a single-size selection step : After adding the appropriate volume of Bead Suspension to the DNA sample, beads will bind larger fragments. The supernatant contains smaller fragments and contaminants that are discarded. For most NGS sequencing applications it is optimal to remove all fragments below ca. 200 bp. This can be achieved by using a volume ratio (bead suspension to sample) of 1.5, which is described in the following protocol.

Before starting
-
Prepare
50mLfresh 80% Molecular Biology grade Ethanol for MN beads (or 70% for AMPure XP beads). -
Remove the NucleoMag® NGS Bead Suspension (or Beckman Ampure XP) from the fridge. Let for approximately 30 min to bring the bead suspension to room temperature.
-
Then, vortex this Bead Suspension stock solution carefully until homogeneization and fill all wells of a 8-tube PCR strip with this solution, or use a reagent reservoir.
-
Add
60µLof well-homogeneized bead suspension into the first 4 columns of a new PCR microplate with a multichannel pipette. -
Spin down the plate containing the digested-ligated products. Fill each well of a 8-PCR tube strip with Tris-Hcl pH8 10mM.
-
In the DNA digestion plate, add
5µLof Tris 10mM with a multichannel pipette, to be sure that you will be able to take 40µl of digested-ligated DNA.
Binding
This step binds DNA fragments 200bp and larger to the magnetic beads.
- For the first 4 columns of the digestion-ligation plate, Transfer
40µLof the DNA samples in the first 4 columns of the purification plate containing beads and carefully mix by pipetting up and down 10 times. - Incubate the purification plate at room temperature for 5 min.
Separation
- Place the purification plate onto the 96-well magnetic separator.
- Wait at least
0h 5m 0suntil all the beads have been attracted by the magnets or until the liquid appears clear. - The supernatant contains unwanted low molecular weight contaminants and unwanted smaller DNA fragments.
- Remove and discard the supernatant (ca 100 µl) by pipetting.
- Do not disturb the attracted beads while aspirating the supernatant. Remove the supernatant with the multichannel from the opposite side of the well.
1st wash with 80 % ethanol
- Place 80% (or 70%) ethanol in a 25ml reservoir.
- With a multichannel pipette, dispense
200µL80% (or 70%) ethanol into the purification plate without disturbing the bead pellet. - Incubate the purification plate at room temperature for at least 30 s.
- Carefully and completely remove and discard ethanol by pipetting.
2nd wash with 80 % ethanol
- With a multichannel pipette, dispense
200µL80% (or 70%) ethanol into the purification plate without disturbing the bead pellet. - Incubate the purification plate at room temperature for at least 30 s.
- Carefully remove and discard ethanol by pipetting.
Dry the beads
Let the purification plate on the magnetic separator and incubate at room temperature for maximum 5 min in order to allow the remaining traces of ethanol to evaporate.
NOTE: take care not to over dry the bead pellet (bead pellet appears cracked in this case) as this will significantly decrease elution efficiency.
Elute DNA fragments library
- Take the purificaation plate from the magnetic stand, and add
40µLof Tris 10mM pH 8 with a multichannel pipette to resuspend the bead pellet by pipetting up and down 10 times. - Incubate the purification plate at room temperature for
0h 5m 0s. - Separate the magnetic beads against the side of the wells by placing the 96-well plate on the magnetic separator.
- Wait at least
0h 5m 0suntil all the beads have been attracted by the magnets or until the liquid appears clear. - Transfer
35µLof the supernatant containing the purified DNA fragments to a new 96‑well plate. Be careful to avoid pipeting beads during this step. - Store the plate at 4°C until and proceed to purification of columns 5-8 and 9-12 similarly (steps 11-15).
- Seal the plate and store at 4°C (or store -20°C for a long-term storage) until PCR amplification.
PCR amplification to generate Illumina sequencing indexed libraries
In this PCR, Illumina indexed primers are incorporated in order to produce fragments compatible with Illumina sequencing, and to insert an index allowing multiplexing of barcoded samples (see ligation step for details). This index will be read during the sequencing run.
This PCR is expected to have a homogenizing effect. Primers are thus included in limiting quantity, in order to produce equalized amounts of PCR fragments among samples. The number of cycles is limited to a maximum of 15 (optimal with 12). After those cycles, a final PCR cycle is then performed after addition of primers in large excess.
The Reaction mixture for a total volume of 40μL is :
| A | B | C | D |
|---|---|---|---|
| conc initiale | conc final | vol 1X µl | |
| Q5 buffer NEB | 5X | 1x | 8 |
| dNTP mix | 25mM each | 0.2 mM | 0.3 |
| primer mix (illPCR1 and illPCR2index) | 5µM each | 0.17 µM | 1.4 |
| Q5 hotstart hifi DNA polymerase NEB | 2u/µl | 0.4 | |
| High GC enhancer NEB | 5X | 1X | 8 |
| nuclease-free water | 11.9 | ||
| Adaptor-ligated purified template DNA | 10 | ||
| total mix | 30 | ||
| TOTAL reaction | 40 |
PCR mixture composition
1st mix preparation
Defreeze and vortex all reagents, except enzymes (stored at -20°C), for approximately 0h 0m 5s
Spin down all reagents for approximately 0h 0m 5s and place on ice.
In a 5ml microtube, prepare the 1st mix according to the following table :
| A | B | C |
|---|---|---|
| Vol µl 1X µl | Vol µl for 1 plate (104 reactions) | |
| Q5 buffer 5X | 8 | 832 |
| dNTP mix 25mM | 0.3 | 31.2 |
| Q5 hot start DNA polymerase 2u/µl | 0.4 | 41.6 |
| High GC enhancer 5X | 8 | 832 |
| nuclease free water | 3.3 | 343.2 |
| total | 20 | 2080 |
First mix composition
Vortex mix all reagents in the mix and spin down.
Aliquot 130µL of the 1st mix into each well of a 8-tube PCR strip.
In a new PCR plate , dispense 20µL of 1st mix with a multichannel in each column.
2nd mix preparation (primer PCR mix) :
prepare 1 mix for each index in a 1.5 ml microtube, (4 mix for a full a plate).
for 1 index
| A | B | C |
|---|---|---|
| vol 1x µl | Vol µl for 3 columns | |
| illPCR Primer mix 5µM | 1.4 | 39.2 |
| nuclease-free water | 8.6 | 240.8 |
| total | 10 | 280 |
Second mix composition (primer PCR mix)
Vortex mix all reagents and spin down.
In a 12-tubes PCR strip, aliquot 93µL of the primer PCR mix into 3 consecutive wells according to the following scheme :

In the PCR plate , dispense 10µL of 2nd mix in each line of the plate with a 12 multichannel pipette, onto the 20µl of the first mix.
DNA and mix combination
Spin down the ligation purified DNA plate.
With a multichannel pipette, transfer 10µL of purified ligated DNA into the PCR plate and mix by pipetting.
Finally, aliquot the 40µL by dispensing 10µL into 3 additional new empty PCR plates.
Seal the 4 PCR plates and spin down.
The 4 PCR will be performed in parallel In 4 different thermal cyclers, in order to reduce the PCR bias.
| A | B | C | D |
|---|---|---|---|
| hot start initial denaturation | 98°C | 30s | |
| amplification cycles | |||
| denaturation | 98°C | 20s | |
| annealing | 60°C | 60s | 15 cycles |
| extension | 72°C | 40s | |
| final extension | 72°C | 10min | |
| hold | 12°C |
PCR program for the Illumina indexing PCR
Seal the 4 PCR plates and spin down.
The 4 PCR will be performed in parallel In 4 different thermal cyclers, in order to reduce the PCR bias.
After PCR, pool back the 4 PCR plates into a single plate with a multichanel.
Final cycle (with pirmers in large excess)
Prepare 1 mix for each index in a 1.5 ml microtube
| A | B | C | D | E |
|---|---|---|---|---|
| conc stock | final concentration | vol µl 1X | vol µl for 3 columns | |
| Q5 buffer | 5X | 1X | 0.2 | 24 |
| dNTP mix | 25mM each | 0.2mM | 0.08 | 9.6 |
| illPCR primer mix | 5µM each | 0.2µM | 0.4 | 48 |
| nuclease-free water | 0.32 | 38.4 | ||
| total | 4 | 120 |
Vortex mix all reagents and spin down. In a 12-tube PCR strip, aliquot 40µL of the mix into 3 consecutive wells according to the following scheme:

Dispense4µL of final cycle mix in each line of the plate with a 12 multichannel pipette.
Seal the PCR plate and spin down.
In a thermocycler, run the final cycle as follows:
| A | B | C |
|---|---|---|
| denaturing | 98°C | 3 min |
| annealing | 60°C | 2 min |
| extension | 72°C | 12 min |
| hold | 12°C |
PCR program for the final cycle of the illumina PCR
Place at 4°C (or -20°C for a long-term storage).
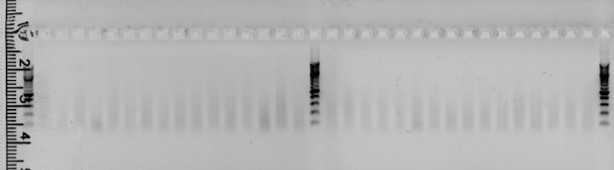
Agarose gel electrophoresis
Check the efficiency of the PCR on an agarose 1.5% gel . Load 5µl of each PCR product.
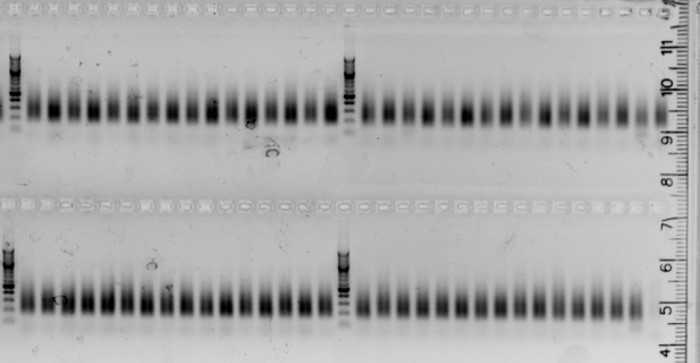
Samples pooling
Each barcoded and indexed individual can now be pooled in a single tube, in equimolar conditions.
After the normalizing PCR, all smears should have similar intensity on the agarose gel. In this case, pool 5 µl of all individuals in a single low binding 1.5 ml microtube.
If not, normalization can be made at this step. For this, roughly estimate the concentration of fragments from the gel picture, and pools accordingly. It can be efficient to make intermediate pools (for example, one pool for the low, one for the medium, and another one for the high intensity samples. Proceed to next step.
Estimate the double strand DNA concentration in the pool by fluorimetry with a Qubit equipment. If the concentration is too low (less than 20 nM), perform a bead clean-up (ratio 1:1, see above steps 9-16) on a partial volume of the pool to increase the final concentration. Work in a microtube instead of a microplate, with a magnetic microtube stand. Adapt the elution volume to the desired final concentration. Elution should be done in at least 50µl, for subsequent steps.
Size selection with sage science Pippin-Prep
Perform the size selection of fragments between 300 and 800 pb using a 1,5% DF marker K agarose gel cassette, according to the Pippin prep manufacturer's instructions :
Quick-Guide-CDF1510-marker-K3.pdf
Warning :
If you perform the size selection on the pool without prior bead clean-up, you may observe a shift between observed and expected size ranges of the smear (approximately 250-750 obtained instead of 300-800).
If you see adaptor dimers on the gel after PCR, it is also preferable to perform a bead clean-up before size selection in the Pippin Prep.
In the case you do not have access to a Pippin prep, you can alternatively perform a double size-selection with beads (see the bead manufacturer's instructions for details), or by smear excision and purification from an agarose gel. From our experience, size selection with the Pippin prep is the most accurate and repeatable method, and has to be prefered if you have serveral pools of libraries (for example 3 pools of 192 samples) in order to increase the number of common loci between samples in the ddRAD sub sampling of the genome.
Quality control of the libraries
Control the quality of the library with a Bioanalyzer (Agilent) in a High Sensitivity DNA chip. Dilute your pool 1:2 or more and load 1µl of the pool before and after size selection, according to the manufacturer's instructions :
HighSensitivity_DNA_KG.pdf.pdf
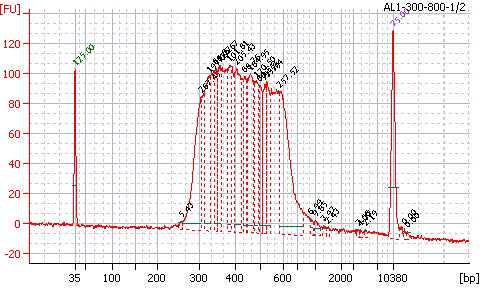
Fluorimetric estimation of the dsDNA concentration in the library.
Perform a quick estimation of the DNA concentration with a fluorimetric assay, in a QubitTM apparatus or equivalent, with the Qubit ds 1X DNA BR assay kit, according to the manufacturer's instructions:
In the case you need an accurate estimation of the DNA concentration in your library, perform a qPCR quantification with the NEBNext Library Quant Kit for Illumina, or equivalent, which uses P5 and P7 illumina primers to target the double stranded DNA fragments in the library. Follows the kit's user guide and perform your quantitative qPCR in a qPCR thermocycler (e.g. LightCycler 480, Roche).
Contrarily to the fluorimetric method (Qubit), the qPCR estimation will only consider dsDNA fragments starting with P5 and ending with P7 illumina sequences, that will be effectively amplified onto the flowcell of the Illumina sequencer.
Suggestions to prepare library dilutions :
in a 8-tube PCR strip:
Prepare intermediate dilutions (1:10 and 1:100) of the library with the dilution buffer supplied in the qPCR kit.
Then prepare the 4 library dilutions to be used in triplicate for qPCR analysis :
1:1000 : 10µl of 1/100 + 90µl buffer 1X
1:2000 : 50µl of 1/1000 + 50µl buffer 1X
1:4000 : 50µl of 1/2000 + 50µl buffer 1X
1:8000 : 50µl of 1/4000 + 50µl buffer 1X
You should get more than 10 nM, that is the library concentration usually required by the sequencing platform facilities.
The library is now ready for sequencing in single read or paired-end 150 bases in an Illumina sequencer, with one index read. Use the average size of the library size range as estimated from the Bioanalyzer profile to convert DNA concentration from nM to ng/µl.