Confirmation of axenic seedlings
Elena L. Peredo, Suzanne M Thomas, Zoe Cardon
Abstract
Sporobolus alterniflorus plants are known to be rich in endophytes. In order to ensure the success of an in vitro culturing approach, it is crucial to remove any potential contaminants. Here, we present a protocol for detecting the presence of endophytes in in vitro plants. To obtain a comprehensive assessment of the culture's axenic state, we employ both culture-dependent and culture-free approaches.
Before start
Please consult your institution's guidelines regarding the culturing and discarding of microorganisms isolated from natural environments.
Steps
Collection of plant materials to be tested for presence of bacteria and/or fungi
The aim of this protocol is to test for the presence of contaminants in putative axenic Sporobolus alterniflorus (smooth cordgrass) seedlings. Precautions should be taken to prevent the in vitro plant material ("vitroplants") from getting exposed to secondary sources of contamination during testing, including preparing the plant materials in a tissue culture hood and using autoclaved supplies.
Prepare plant materials.
We are going to use two different procedures for testing for the presence of bacteria and fungi in our cultures. We will prepare both simultaneously.
- Prepare two racks with labeled, sterile 1.7 mL Eppendorf tubes. Samples in one rack would be used for culture-dependent methods. The second rack is for culture-independent methods.
- Place in each tube the plant material to be tested. Use autoclaved scissors to cut fragments of around 2-3 cm of leaves and roots.
Culture-dependent methods
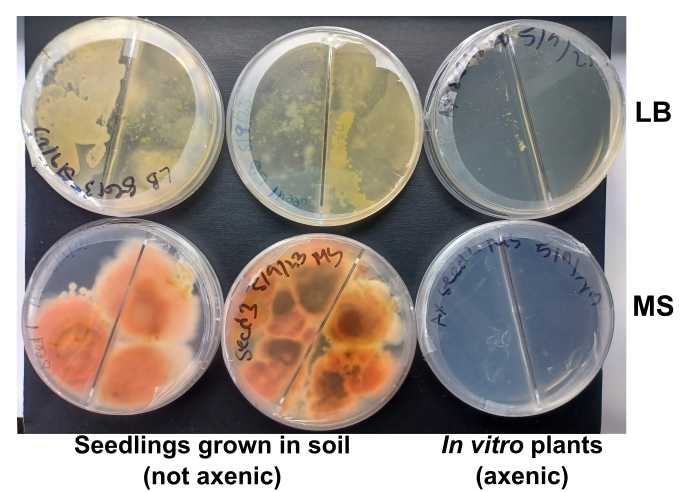
We use here two different culture media to detect the presence of possible contaminants. One of the media is the same medium the plants are growing in. The second is a rich, general-purpose medium broadly used in microbiology.
Prepare culture media.
Lysogenic Broth culture medium (LB)
- Weigh 25g of LB broth powder and add to a 1L autoclave bottle.
- Add double distilled water for a final volume of 1L.
- Adjust pH to ~7.
- Add 10 g/L of agar.
- Autoclave for 40 minutes.
- In the hood, add ~25 mL into as many 90 mm sterile Petri dishes as needed.
Plant culture medium (MS)
This culture medium is a modification of Murashige and Skoog (1962) and can be bought as dry powder from Phytotech Labs (reference M527).
-
Add 4.41 g of powder to a 1L autoclave bottle.
-
Add 30 g/L of sucrose for a final concentration of 3%.
-
Add double distilled water for a final volume of 1L.
-
Adjust the pH to 5.8.
-
Add 8 g/L of agar.
-
Autoclave for 40 minutes. In the hood, add ~25 mL into as many 90 mm sterile Petri dishes as needed.
Prepare plant extracts
- Add 500 µL of distilled autoclaved water (or molecular-grade water) to each sterile Eppendorf tube.
- Use a sterile micro-pestle to grind the plant material to a fine paste. (Alternatively, a bead-beating approach can be used.)
- Transfer 100 µL of the supernatant to the appropriate media for culture.
- Use a glass spreader to distribute the sample on the surface of the agar.
- Incubate for 1 week using a temperature and light regimen similar to that used for growing seedlings. (We use a growth chamber temperature of 20$$°C, light level ~100 µE, and 16 hr: 8 hr light:dark cycles.)
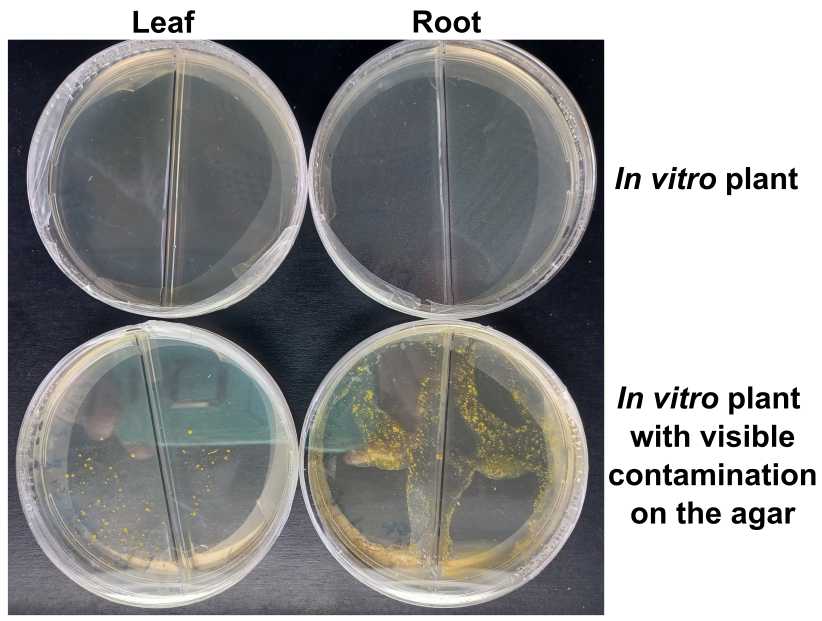
Score the presence of bacterial of fungal contamination in the culturing plates
An example:
Culture-independent methods
We use a PCR-based detection method with universal primers targeting the 16S rRNA gene to detect the possible presence of endogenous bacteria. This second method complements the culture-depended assay described above, targeting bacteria that might not be suitable to grow in the culture media defined above but that can still be present in the in vitro plants.
In addition to the standard negative controls, we strongly encourage using positive control for amplification including
- Bacterial DNA (e.g. DNA from a bacterial isolate).
- Positive control for bacterial contamination of plant material (e.g. soil-grown seedlings, or plants collected in the field).
DNA extraction
- Add the appropriate volume (µL) of DNA extraction buffer (e.g. CTAB) to each tube containing the plant material to be evaluated using the PCR-based assay.
- Use a sterile micro-pestle to grind the plant material to a fine paste. (Alternatively, a bead-beating approach can be used.)
- Follow the instructions provided by manufacturer for your DNA extraction kit of choice (here we used Maxwell® RSC PureFood GMO and Authentication Kit).
- Determine the DNA quantity and quality using nanodrop, and/or agarose gel, and/or fluorometric quantification.
- Prepare working solutions of each DNA (~10 ng/µL).
Primers for the amplification of the 16S rRNA gene
We recommend using the primer set 799F-1192R targeting the 16S rRNA gene (Ma et al., 2022). This primer combination has the advantage of a reduced affinity for the 16S rRNA gene in cyanobacteria, and therefore minimizes the amplification of chloroplast DNA.
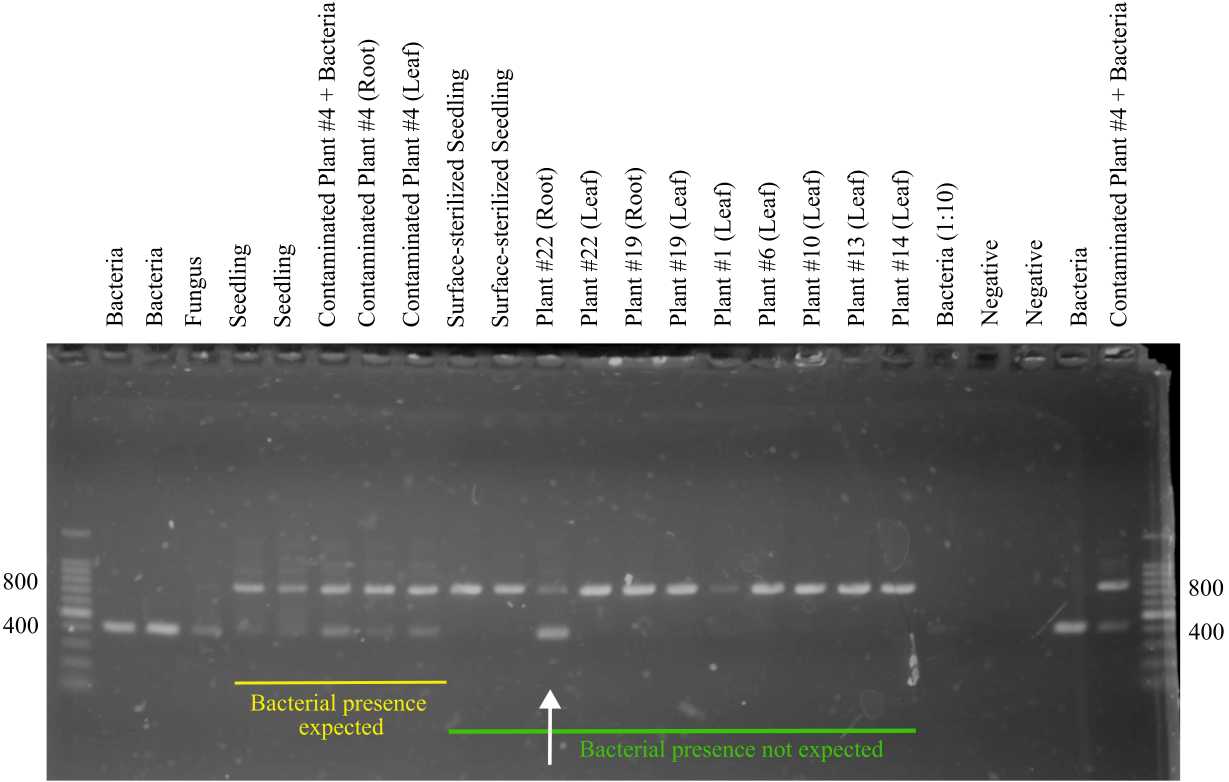
This primer combination amplifies a ~425 bp fragment of the 16S rRNA gene in bacteria. In Sporobolus alterniflorus it also amplifies a ~800 bp fragment, corresponding to the 16S rRNA gene in the plant mitochondrial genome.
| A | B |
|---|---|
| 799F | AACMGGATTAGATACCCKG |
| 1192R | ACGTCATCCCCACCTTCC |
| 27F | AGAGTTTGATCCTGGCTCAG |
| 1492R | TACGGYTACCTTGTTACGACTT |
Table 1. Primers used for amplification of 16S gene.
PCR reaction
We selected Platinum™ SuperFi™ DNA Polymerase (Invitrogen) as the enzyme in our PCR reactions. Many other Taqs will provide successful results, however, we recommend using a high-fidelity, high-efficiency enzyme in this protocol.
| A | B | C |
|---|---|---|
| Buffer | 5X | 4 |
| DNA | 10 µg/µL | 2 |
| dNTPs | 2.5 mM | 0.5 |
| Primer F | 10 µM | 1 |
| Primer R | 10 µM | 1 |
| Taq | 2 U/μL | 0.2 |
| Water | 9.5 |
Table 2. Components of the PCR reaction. The final volume per reaction is 20 µL.
PCR conditions
- Initial denaturation at 94°C for 5 min.
- Ten cycles of a touchdown PCR. In each cycle, we decreased the annealing temperature by 0.5°C. Each cycle consisted of denaturation at 94°C for 30 sec, followed by annealing at 60°C for 30 sec, and extension at 72°C for 45 sec.
- Twenty-five cycles of denaturation at 94°C for 30 sec, followed by annealing at 55°C for 30 sec, and extension at 72°C for 45 sec.
- Final elogation at 72°C for 5 min.
- Samples were held at 4°C.
Agarose detection of PCR fragments
The expected sizes of the fragments produced in this reaction by the primer set 799F/1199R are ~400 bp (bacteria) and ~800 bp ( Sporobolus alterniflorus mitochondria).
- Prepare a 1% agarose gel, you can use TAE 1x as the electrophoresis buffer.
- Add SyBr to the agarose, for a final concentration 1x, following manufacturer instructions.
- For each PCR reaction, separate 10 µL of PCR product and mix with 2 uL of loading dye.
- Load each sample in the agarose gel.
- Use a 1 Kb ladder (or equivalent) as a size marker (follow manufacturer instructions).
- Run the until the bands separate. We run our gels at 100 v for 1.5-2h but these conditions will depend on the length of the gel.
- Visualize the results of the gel in a gel reader.
Sequencing
Another way of visualizing bacterial contamination in a given sample is by visual inspection of the Sanger sequencing electropherograms.
- Samples with bacterial contamination will display low-quality base-calling results up to position ~425 due to the presence of a secondary band that corresponds to the bacterial 16S rRNA gene. The rest of the electropherogram, corresponding to the sequence of the mitochondria 16s ribosomic gene, will show as high-quality base calls.
- Samples corresponding to axenic vitroplants will display sequences of >700 bp with high-quality results for all the bases, consistent with the amplification of a single target gene (the mitochondria 16S ribosomic gene).
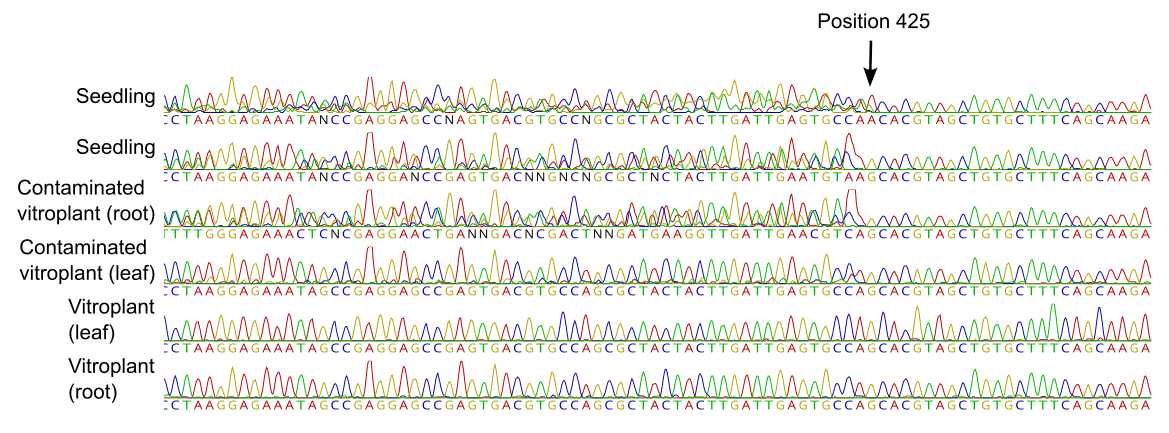
Fig. 4- Sanger sequencing electropherograms.

