Co-extraction of RNA and DNA from soil and sediment samples
Dominik Buchner
Abstract
This protocol describes extracting RNA and DNA from soil and sediment samples. 2g of soil or up to 5g of sediment can be processed in one extraction. The protocol is based on the RNeasy PowerSoil Total RNA Kit but does not rely on any components of the kit. It also replaces the RNeasy PowerSoil DNA Elution kit. The JetStar 2.0 columns are replaced with the HiPure Columns from Invitrogen's PureLink HiPure Plasmid Miniprep Kit. A lot of the buffers can be found in the following patent https://patents.google.com/patent/US7459548B2/en.
Before start
Make sure all buffers are prepared before starting.
Steps
Cell lysis
Prepare one 15 mL conical centrifuge tube per sample by adding 0.5 g of a) 0.1 mm glass beads, b) 0.5 mm glass beads, c) 1 mm zirconia/silica beads d) 2 mm zirconia beads.
Control that the cap is closed properly. Vortex until the biphasic layer disappears.
2500x g
Transfer 3.5mL of the upper aqueous phase to a new 15 mL centrifuge tube. Make sure not to pierce the biphasic layer and to not transfer any liquid from the lower phenol phase. If this happens by accident, recentrifuge the sample and repeat. In soil high in organic matter the biphasic layer will be much thicker than in sediment samples.
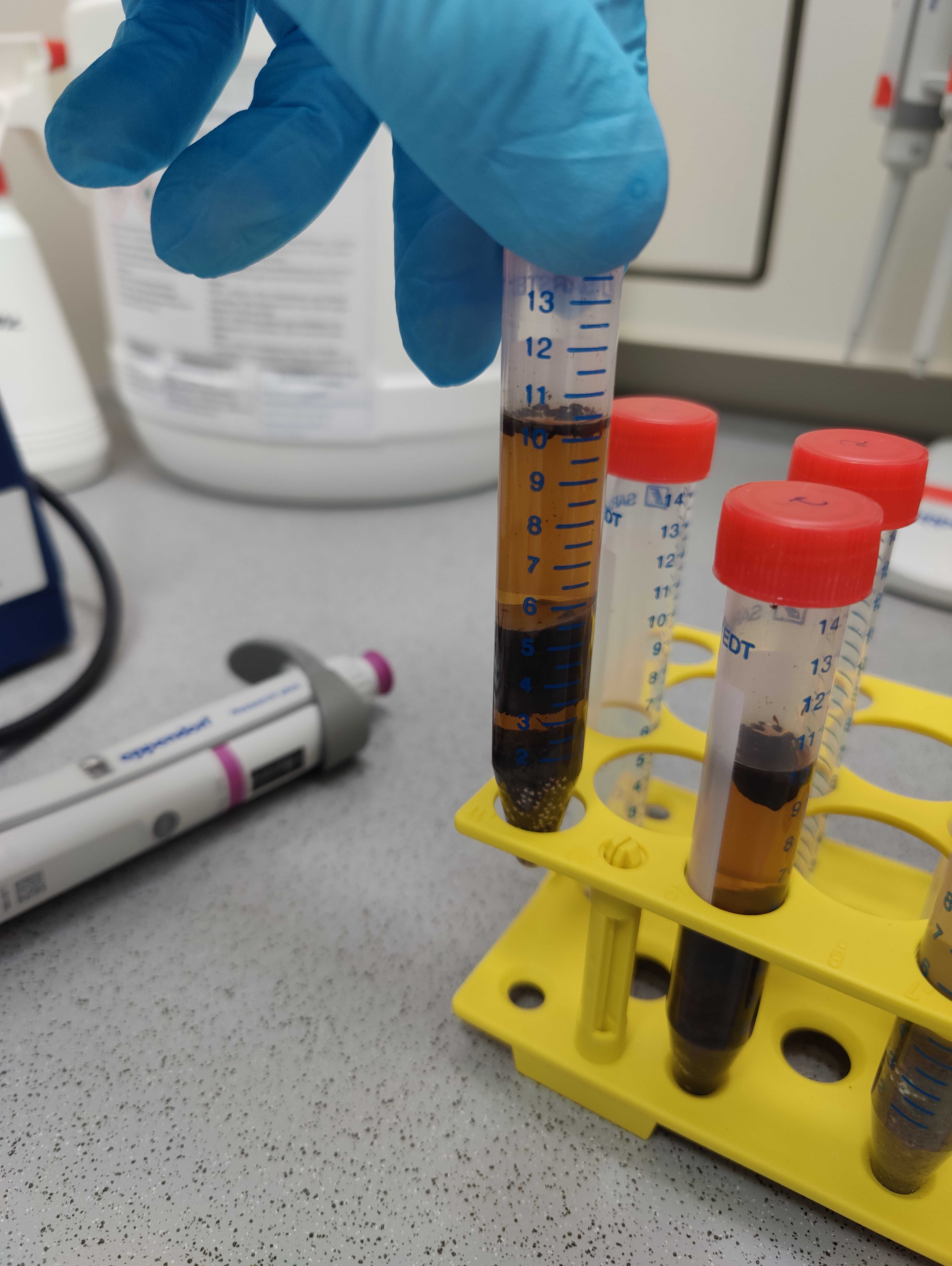

Inhibitor removal
Incubate at 4°C for 0h 10m 0s .
2500x g
Without disturbing the pellet (if there is one), transfer the supernatant to a new 15 mL centrifuge tube.
Nucleic acid precipitation
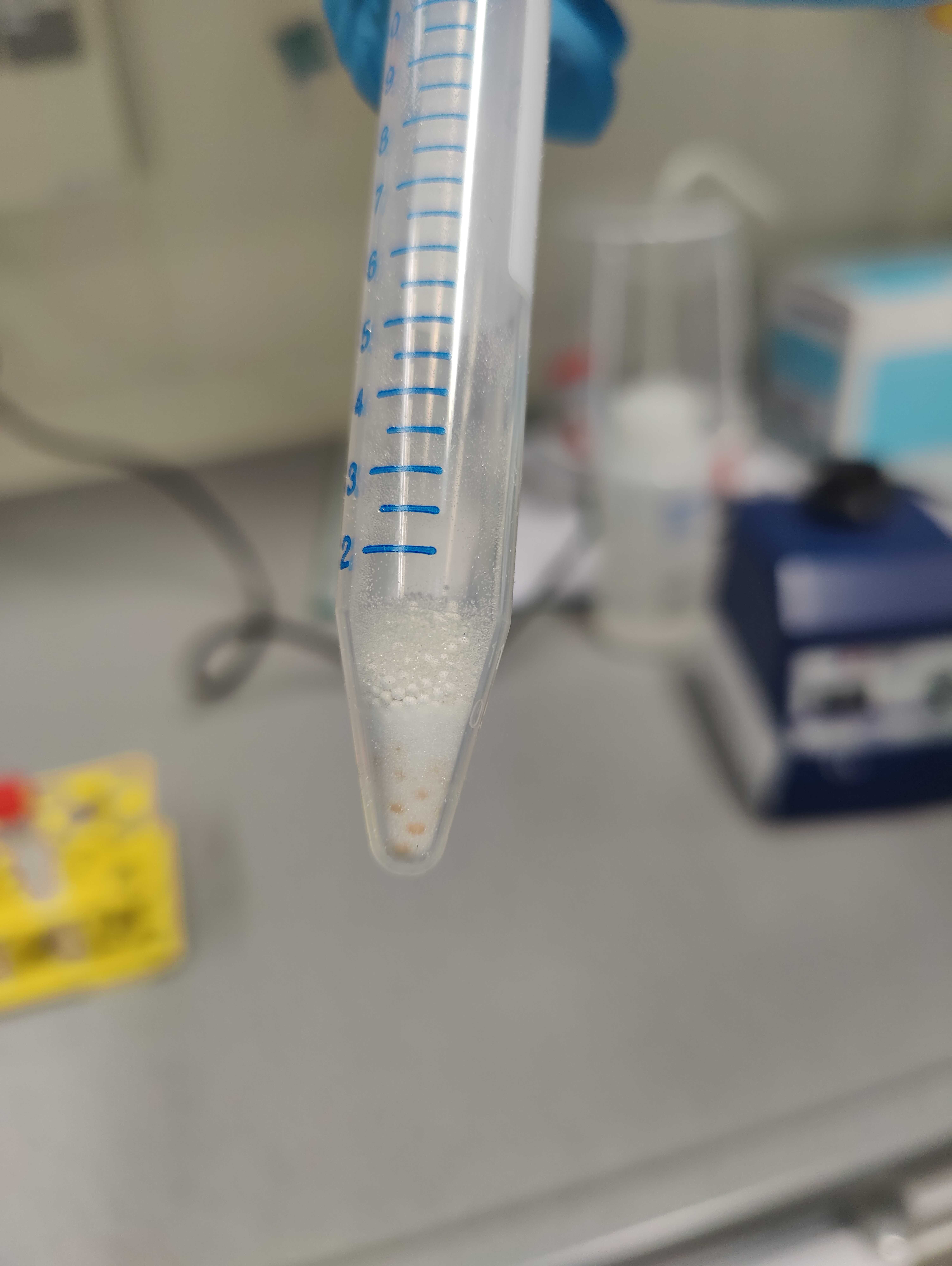
Add 5mL isopropanol. Mix by vortexing and incubate at Room temperature for 0h 30m 0s .
2500x g .
Anion exchange clean-up
Add 1mL to the pellet and incubate the sample at 45°C for 0h 10m 0s . Afterwards, dissolve the pellet by vortexing and/or pipetting.
Place a HiPure or JetStar 2.0 column in a 15 mL centrifuge tube or a suitable rack.

Equilibrate the column with 2mL . Wait until all of the buffer volume has passed the column by gravity flow.
Load the sample from step 15 on the equilibrated column. Make sure all that the pellet is dissolved properly or it might heavily reduce the flow-through and reduce the expected yield. Wait until all of the buffer volume has passed the column by gravity flow.
Wash the column with 1mL . Wait until all of the buffer volume has passed the column by gravity flow.
Transfer the column to a new 15 mL centrifuge tube or place a 2 mL collection tube in the rack to collect the RNA.
Add 1mL to the column. Wait until all of the buffer volume has passed the column by gravity flow.
Transfer the column to a new 15 mL centrifuge tube or place a new 2 mL collection tube in the rack to collect the DNA
Add 1mL to the column. Wait until all of the buffer volume has passed the column by gravity flow.
If a 15 mL centrifuge tube was used for elution, transfer the DNA/RNA sample to a 2 mL tube.
Add 1 mL of isopropanol and incubate at -20°C for 0h 15m 0s
13.000x g
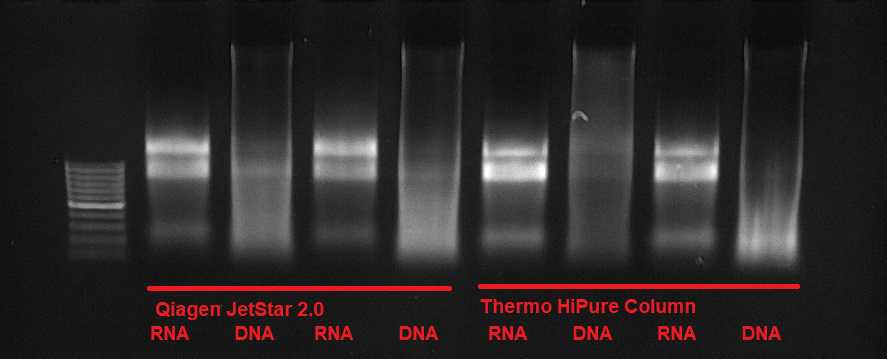
Resuspend the DNA/RNA in 100µL . Check the integrity of the DNA/RNA on an agarose gel. afterward