Cell-Free Protein Synthesis
Lena Thoring, Stefan Kubick, Anne Zemella, Theresa Richter
Cell-free protein synthesis
G protein-coupled receptor
Protein modification
Non-canonical amino acids
Amber suppression
Confocal laser scanning microscopy
Abstract
This is part 3.3 of the "A Combined Cell-Free Protein Synthesis and Fluorescence-Based Approach to Investigate GPCR Binding Properties" collection of protocols : https://www.protocols.io/view/a-combined-cell-free-protein-synthesis-and-fluores-bqntmven https://www.protocols.io/view/a-combined-cell-free-protein-synthesis-and-fluores-bqntmven
Collection Abstract: Fluorescent labeling of de novo synthesized proteins is in particular a valuable tool for functional and structural studies of membrane proteins. In this context, we present two methods for the site-specific fluorescent labeling of difficult-to-express membrane proteins in combination with cell-free protein synthesis. The cell-free protein synthesis system is based on Chinese Hamster Ovary Cells (CHO) since this system contains endogenous membrane structures derived from the endoplasmic reticulum. These so-called microsomes enable a direct integration of membrane proteins into a biological membrane. In this protocol the first part describes the fluorescent labeling by using a precharged tRNA, loaded with a fluorescent amino acid. The second part describes the preparation of a modified aminoacyl-tRNA-synthetase and a suppressor tRNA that are applied to the CHO cell-free system to enable the incorporation of a non-canonical amino acid. The reactive group of the non-canonical amino acid is further coupled to a fluorescent dye. Both methods utilize the amber stop codon suppression technology. The successful fluorescent labeling of the model G protein-coupled receptor adenosine A2A (Adora2a) is analyzed by in-gel-fluorescence, a reporter protein assay, and confocal laser scanning microscopy (CLSM). Moreover, a ligand-dependent conformational change of the fluorescently labeled Adora2a was analyzed by bioluminescence resonance energy transfer (BRET).
For Introduction and Notes , please see: https://www.protocols.io/view/a-combined-cell-free-protein-synthesis-and-fluores-bqntmven/guidelines
Steps
3.3 Cell-Free Protein Synthesis
Thaw all required components for cell-free protein synthesis On ice ( see Note 8 ).
The cell-free protein synthesis is performed in a coupled mode where transcription and translation reaction take place in one vessel. A standard reaction is composed of 40%, 10micromolar (µM), 1x, and 1x. Add plasmid at a concentration of 40nanomolar (nM) ( see Note 9 ). Add 1U/μl for the transcription reaction and 14C-leucine (specific radioactivity of 66.67 dpm/pmol) for further analysis of de novo synthesized proteins. Fill the reaction with water to the final volume of 50µL.
3.3.1 Fluorescent Labeling with Bodipy-TMR-Lysine
Thaw the Bodipy-TMR-lysine-tRNACUA (BP-CUA) and Bodipy-TMR-lysine-tRNAGAA (BP-GAA) On ice and keep it dark ( see Note 10 ).
Pipette 2micromolar (µM) to the cell-free reaction and incubate the prepared cell-free protein synthesis reaction by 27°C for 3h 0m 0s and shaking at 600rpm ( see Note 11 ). Cover the thermomixer with a lid or aluminum foil to prevent the reaction of any light.
Take a 5µL aliquot of the translation mixture for SDS-PAGE. Centrifuge the translation reaction at 16000x g,4°C. Take 5µL of the supernatant and resuspend the pellet (microsomal fraction) in an equal volume PBS in comparison to the volume of the translation reaction. This step is required for the analysis of the localization of the synthesized protein. The expectations of the incorporation of the different Bodipy-tRNAs are described in Table 1 ( see Note 12 ).
| A | B | C | D |
|---|---|---|---|
| Construct | Precharged tRNA | Expectation | |
| Synthesis | Fluorescence | ||
| Adora2a | BP-GAA | Full-length protein | Highly fluorescently labeled protein with |
| statistical incorporation of ncAA | |||
| Adora2a | BP-CUA | Full-length protein | The full-length protein is not labeled with the |
| fluorescent dye and the ncAA is not | |||
| incorporated. | |||
| Adora2a_amb | BP-CUA | Full-length protein and | |
| partial termination | |||
| product | The full-length protein is fluorescently labeled. | ||
| The fluorescence signal has a lower intensity | |||
| in comparison to the statistically labeled | |||
| Adora2a due to the site-specific incorporation | |||
| of the ncAA at one defined position. | |||
| Termination product is not labeled. | |||
| Adora2a_amb | BP-GAA | Termination product | The termination product is highly fluorescently |
| labeled due to the statistical incorporation of | |||
| the ncAA. No full-length product is visible. |
Table 1Expectations of cell-free protein synthesis and fluorescent labeling of different Adora2a constructs with Bodipy-tRNA directed to phenylalanine codons (GAA) or directed to the amber stop codon (CUA)
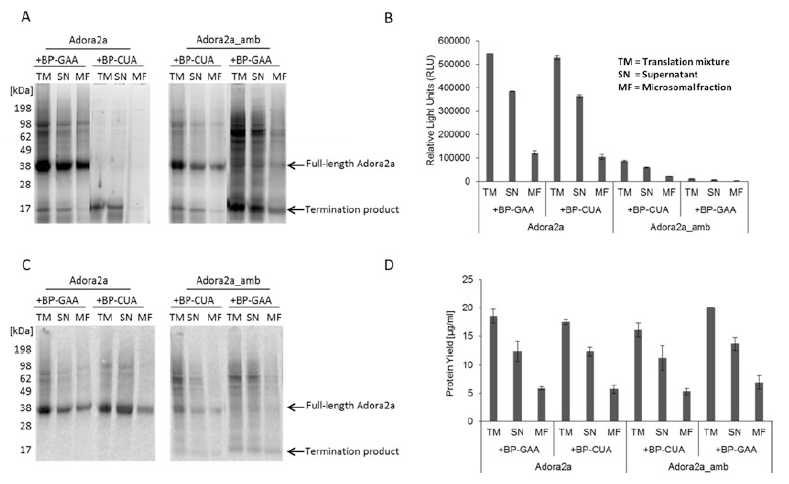
An example of the performance of the fluorescent labeling of Adora2a with or without incorporated non-canonical amino acid determined by in-gel-fluorescence and autoradiography, and the measurement of the reporter protein are shown in Fig. 2. In-gel-fluorescence and autoradiography (Fig. 2a, c) showed the expected results as described in Table 1. The reporter protein assay (Fig. 2b) is based on the activity of a receptor C-terminally fused Nanoluciferase (Nluc). Nluc activity can only be measured after the translation of the full-length fusion protein. The Adora2a synthesized by using the DNA construct without an amber stop codon (Adora2a + BP-GAA/ BP-CUA) resulted in a fusion protein with a detectable Nluc activity. The fusion protein translated from the amber stop codon DNA construct (Adora2a_amb + BP-CUA) showed a fivefold reduced Nluc activity. This result implicates a rather low incorporation efficiency of the precharged tRNA addressing the amber stop codon. The combination of Adora2a_amb with BP-GAA showed no luciferase activity since only the termination product was translated. Protein yields were calculated by scintillation counting and resulted in approximately 20μg/ml in translation mixture. In the microsomal fraction and the supernatant, approximately 6μg/ml and 14μg/ml were detected, respectively. A similar Adora2a distribution was obtained by in-gel-fluorescence, luciferase activity, and autoradiography. Previously reported distributions of membrane proteins are comparable to the here described distribution [7]. A comparable protein yield was calculated for the termination product.
3.3.2 Analysis of Ligand- Dependent Conformational Change Using a BRET Assay
Site-specifically label the Adora2a_amb using the precharged tRNA Bodipy-TMR-lysine-tRNACUA as described in section 3.3.1 above, steps 3–5.
Resuspend the microsomal fraction of the Adora2a_amb in PBS. 5µL aliquots of resuspended Adora2a_amb were mixed with 5µL in PBS with final concentrations of 0micromolar (µM), 100micromolar (µM), 1000micromolar (µM), and 5000micromolar (µM) ( see Note 13 ).
10µL were applied for the luminescence and fluorescence measurement. In a first step, the luminescence of the Nluc was detected using an OD2 filter. In a second step, the fluorescence of the coupled Bodipy dye, excited by the Nluc emission was detected.
As a control of the resuspended microsomal fraction of 5µL of the resuspended microsomal fraction of the full-length Adora2a protein without any fluorescent label is treated with the same concentrations of adenosine ( step 8 ) to determine background fluorescence caused by the broad emission spectrum of the Nluc and possible interactions of adenosine with the Nluc.
The BRET ratio is calculated as follows:
Herein, the calculated BRET ratio (Fig. 3) showed a change in the relation of the fluorescence of the Bodipy in comparison to the Nluc luminescence after the addition of different adenosine concentrations. Only a minimal increase of the BRET ratio can be seen after addition of higher adenosine concentrations (above 100micromolar (µM)) indicating that at lower concentrations all receptors are occupied with adenosine. The result indicates a conformational change of the helix III and the connected third intracellular loop that is expected for the Adora2a.

3.3.3 Site-Specific Incorporation of a Non-canonical Amino Acid with Subsequent Fluorescent Labeling and Microscopic Analysis
Additional components are required for the recharging of the suppressor tRNA and a subsequent incorporation of a
non-canonical amino acid. Therefore, add the p-propargyloxy-L-phenylalanine, tRNATyrCUA, and eAzFRS in a specific order (Table 2) (see Note 14 ).
| A | B | C |
|---|---|---|
| Order | Components | Final concentration |
| 1 | Water | |
| 2 | PolyG | 10 μM |
| 3 | Translation mix | 1× |
| 4 | pPa | 2 mM |
| 5 | DNA-template | 40 nM |
| 6 | Lysate | 40% |
| 7 | tRNATyrCUA | 3 μM |
| 8 | eAzFRS | 3 μM |
| 9 | 14C-leucine | 66.67 dpm/pmol |
| 10 | T7-RNA-Polymerase | 1 U/μl |
| 11 | Energy mix | 1× |
Table 2Pipetting order of a standard cell-free reaction with orthogonal components for the incorporation of non-canonical amino acids
Incubate the prepared cell-free reaction at 27°C for 3h 0m 0s by gentle shaking at 600rpm. Keep the reaction in the dark ( see Note 15 ).
The membrane protein is translocated and integrated into the microsomal membrane during the cell-free reaction. Therefore, separate the microsomal fraction by centrifugation at 16000x g,4°C. Resuspend the pellet fraction in PBS. Use an equal volume of the cell-free reaction for resuspension.
For the labeling reaction prepare the labeling mix as follows: combine 200micromolar (µM) CuSO4 4 with 600micromolar (µM), 5millimolar (mM), PBS and a final concentration of 3micromolar (µM) to a final volume of 5µL. Add 5µL of the resuspended microsomal fraction to the labeling mix. Incubate the labeling reaction at Room temperature for 1h 0m 0s. Keep the reaction dark ( see Note 16 ).
Centrifuge the labeling reaction for 16000x g,4°C. Discard the supernatant and resuspend the pellet in 10µL. This step removes excess fluorescent dye to decrease the background signal in the subsequent fluorescent analyses.
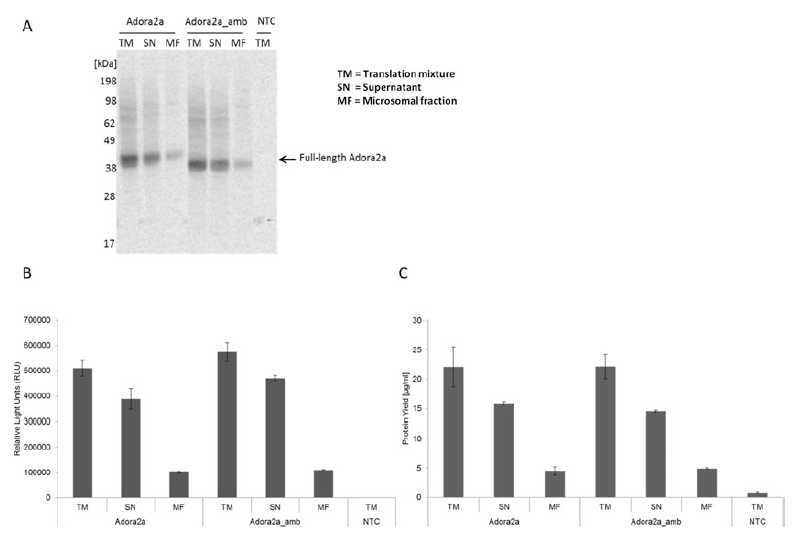
The analysis of the synthesis of the full-length protein by using the Adora2a construct with or without an amber stop codon was performed by autoradiography and a reporter protein assay (Fig. 4). The autoradiography shows for both synthesis reactions a similar band pattern (Fig. 4a). Interestingly, the incorporation of pPa led to a comparable band signal as obtained for the full-length protein translated from the DNA construct without an amber stop codon. This result implicates a high incorporation efficiency of the non-canonical amino acid. In addition, no termination product is detected in the autoradiograph. The high incorporation efficiency is the basis for a further coupling reaction to a fluorescent dye. In addition, this result is supported by the Nluc assay (Fig. 4b). The measured luciferase activity of the suppression product reaches up to 80% of the luciferase activity of the full-length product. The protein yield and protein distribution (Fig. 4c) is comparable to the previously described results (Fig. 2, see section 3.3.1 above "Fluorescent Labeling with Bodipy-TMRLysine", step 6 ).
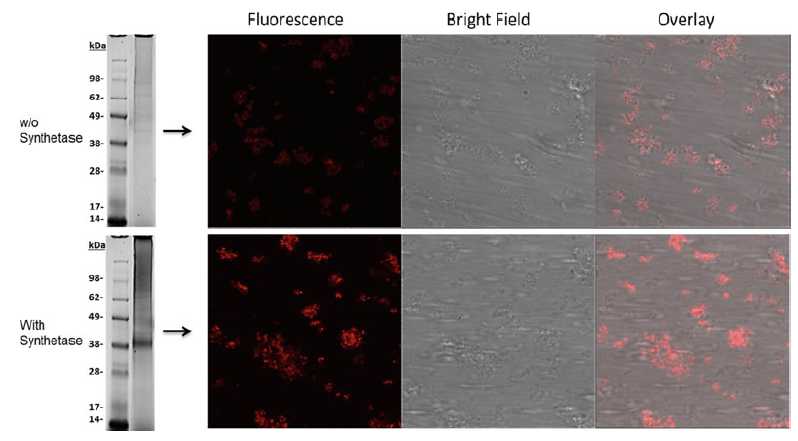
Fluorescently labeled GPCRs should be detectable in microsomal structures (Fig. 5). It is recommended to visualize the labeled sample first by in-gel-fluorescence (Fig. 5, left panel). The synthesis reaction in the presence of all orthogonal components led to a specific band at the expected molecular weight. The control reaction without addition of the modified synthetase resulted in no visible band. The success of the microscopic analysis highly correlates to the quality of the in-gel-fluorescence. High background fluorescence during ingel-analysis often results in unspecific staining of the microsomes. The microscopic analysis clearly shows a difference in the fluorescence intensity of the labeled Adora2a and the unspecific staining of the microsomes (right panel).