Biochemical Separation of Cytoplasmic and Nuclear Fraction for Downstream Molecular Analysis
Hang T. Huynh, Hang T. Huynh, Evgeniia Shcherbinina, Evgeniia Shcherbinina, Hsiang-Chi Huang, Hsiang-Chi Huang, Reyhaneh Rezaei, Reyhaneh Rezaei, Aishe A. Sarshad, Aishe A. Sarshad
biochemical fractionation
cellular compartmentalization
cytoplasmic extraction
nuclear isolation
protein localization
Abstract
Biochemical fractionation is a technique used to isolate and separate distinct cellular compartments, critical for dissecting cellular mechanisms and molecular pathways. Herein we outline a biochemical fraction methodology for isolation of ultra-pure nuclei and cytoplasm. This protocol utilizes hypotonic lysis buffer to suspend cells, coupled with a calibrated centrifugation strategy, for enhanced separation of cytoplasm from the nuclear fraction. Subsequent purification steps ensure the integrity of the isolated nuclear fraction. Overall, this method facilitates accurate protein localization, essential for functional studies, demonstrating its efficacy in separating cellular compartments. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Biochemical fractionation
Support Protocol 1 : Protein quantification using Bradford assay
Support Protocol 2 : SDS/PAGE and Western blotting
OVERVIEW AND PRINCIPLES
The maintenance of cellular homeostasis requires correct protein function. In turn, protein function is directly linked to their subcellular localization. Protein localization influences protein-protein interaction, enzymatic activities, and involvement in signaling pathways. Thus, knowledge of the localization of particular proteins can provide important evidence about its cellular physiology (Jadot et al., 2017).
The mammalian cell is divided into distinct areas where the most profound compartments are the nucleus and the cytosol, which contains other distinct membrane-bound organelles such as the endoplasmic reticulum, mitochondria, and endosomes. In the nucleus DNA transcription, replication and RNA synthesis occurs, while in the cytoplasm protein synthesis takes place (Fu et al., 2018). Nevertheless, the protein distribution inside the cell is dynamic: some proteins can translocate between compartments, i.e., from the cytoplasm to the nucleus, in response to cellular signals or during different stages of the cell cycle. Such movements are crucial for processes like gene expression regulation, cell cycle progression, and response to stress or stimuli. For example, transcription factors such as p53 or NF-Kβ are mainly located in the cytoplasm, but upon receiving an activation signal they translocate into the nucleus and interact with specific DNA loci to mediate the transcription of target genes (Liu et al., 2018). Another protein, Argonaute 2 (AGO2), the main component of RNA interference pathway is usually found within the cytoplasm, yet, in some conditions, such as cell confluence or certain cancers, can relocate to the nucleus (Huang et al., 2024; Johnson et al., 2023; Lobo et al., 2024; Sarshad et al., 2018). Therefore, an appropriate protocol to identify and separate the subcellular protein localization is critical to understand the physiological function of shuttling and localized proteins.
Current methods for subcellular fractionation, which include ultracentrifugation and biochemical fractionation, offer powerful tools for isolating specific cellular compartments. Ultracentrifugation relies on the differential sedimentation rates of cellular components, which vary based on their size, shape, and density, allowing for the isolation of distinct organelles and macromolecular complexes (Alberts, 2002). While ultracentrifugation is invaluable for its ability to separate a wide range of cellular components with remarkable precision, it has few crucial limitations. The technique requires sophisticated and expensive equipment that may not be accessible to all research facilities. Additionally, the high forces involved can potentially damage delicate biological structures, and the process itself can be time-consuming, requiring careful optimization and calibration to achieve the desired separation.
Biochemical fractionation serves as a refined method for separating the cytoplasm from the nucleus in mammalian cells, capitalizing on the differential solubility and density of cellular components when subjected to specific buffer conditions and centrifugation steps (Senichkin et al., 2021). This technique begins with the gentle lysis of cells in a buffer designed to preserve organelle integrity while releasing the soluble cytoplasmic content. The lysate is then subjected to low-speed centrifugation to pellet the nuclei, effectively separating them from the cytoplasmic fraction contained in the supernatant. The principle of the method hinges on the careful control of osmotic conditions and buffer compositions to ensure the selective solubilization of cytoplasmic components, while maintaining nuclear structures intact. One of the significant advantages of biochemical fractionation is its ability to yield highly pure fractions of cytoplasm and nucleus, which allows accurate downstream analysis of proteins and their interactions in each subcellular fraction. Moreover, this approach is less harsh than methods relying on high centrifugal forces, thereby minimizing the risk of organelle damage and preserving the functional integrity of the separated compartments (Alberts, 2002). The adaptability of biochemical fractionation to various cell types and conditions makes it applicable to many studies.
Furthermore, there are commercially available kits for cytoplasmic and nuclear separation. However, these kits are often costly, especially for large scale fractionation, and the reagents and buffer compositions are not disclosed. Therefore, preparing lysis buffers with commonly available reagents in laboratories overcome cost issues and allow for control of the procedure.
Building upon the protocol established by Gagnon et al., herein, we introduce a protocol for the separation of the cytoplasm from the nucleus, which we adjusted to fit many commonly used cell culture (Gagnon et al., 2014). Cells have varying properties i.e., in terms of cell size and stiffness; therefore, it is often suggested to optimize fractionation parameters depending on the cell line that is fractionated. Our approach addresses the limitations of traditional methods by optimizing centrifugation parameters, resulting in higher purity and efficiency of fractionation. We demonstrate that the set of parameters outlined here is suitable for many cancer cell lines used commonly in research labs. These cell lines include, but are not limited to, A375, HEK293, SHSY5Y, SK-N-BE(2) HEPG2, Kelly, HeLa, HAP1, U2OS, Granta-519, Molm-13 and MCF7. This protocol aims to provide a reliable and replicable method for cellular fractionation, facilitating more accurate analyses of protein localization and function.
STRATEGIC PLANNING
While collecting the cell pellet, it is critical to carefully remove all the supernatant. The remaining buffer may change the pH of the hypotonic lysis buffer and cause imbalanced volume between the cytoplasmic and nuclear fraction.
The volume of the buffer for fractionation should be adjusted based on the size of the cell pellet.
All steps should be performed on ice using pre-chilled tubes to minimize protein degradation.
The procedure can be paused after the nuclear pellet is resuspended in nuclear lysis buffer, and before the sonication step, by incubating the lysate at −20°C.
To avoid degradation of the protein lysate by freeze /thaw cycles, divide the lysates into aliquots for downstream application.
STRATEGIC QUESTIONS
Can I use this protocol with frozen pellets? Yes, this is possible, but all liquid should be carefully removed from the pellet before freezing to avoid lysis of the pellet. We recommend freezing down larger pellets, as smaller pellets are more difficult to reconstitute in the hypotonic lysis buffer after defrosting.
SAFETY CONSIDERATIONS
Sonicators are high-frequency sound generators. They are used in this protocol to disrupt nucleic acids. Take caution when using a sonicator. Wear protective ear covers when working with a sonicator and make sure there are no people in the vicinity when a sonicator is turned on.
NOTE : All protocols involving animals must be reviewed and approved by the appropriate Animal Care and Use Committee and must follow regulations for the care and use of laboratory animals. Appropriate informed consent is necessary for obtaining and use of human study material.
Basic Protocol: BIOCHEMICAL FRACTIONATION
Biochemical fractionation is a technique used to isolate and separates distinct cellular compartments. Here we introduce a biochemical fraction methodology for isolation of ultra-pure nuclei and cytoplasm. This protocol utilizes hypotonic lysis buffer to suspend cells, coupled with a calibrated centrifugation strategy for enhanced separation of cytoplasm from the nuclear fraction (Fig. 1). Subsequent purification steps ensure the integrity of the isolated nuclear fraction. After completing the protocol, to check the effectiveness of the fractionation, Western blot should be performed to analyze the localization of compartment specific markers (described in detail in Support Protocol 2). Calreticulin or GAPDH are commonly used as a cytoplasmic marker, while Lamin B1, Lamin A/C or Histone H3 as a nuclear marker. Biochemical fractionation is considered successful when both the cytoplasmic and nuclear fractions are clean and leakage free from unspecific compartment.
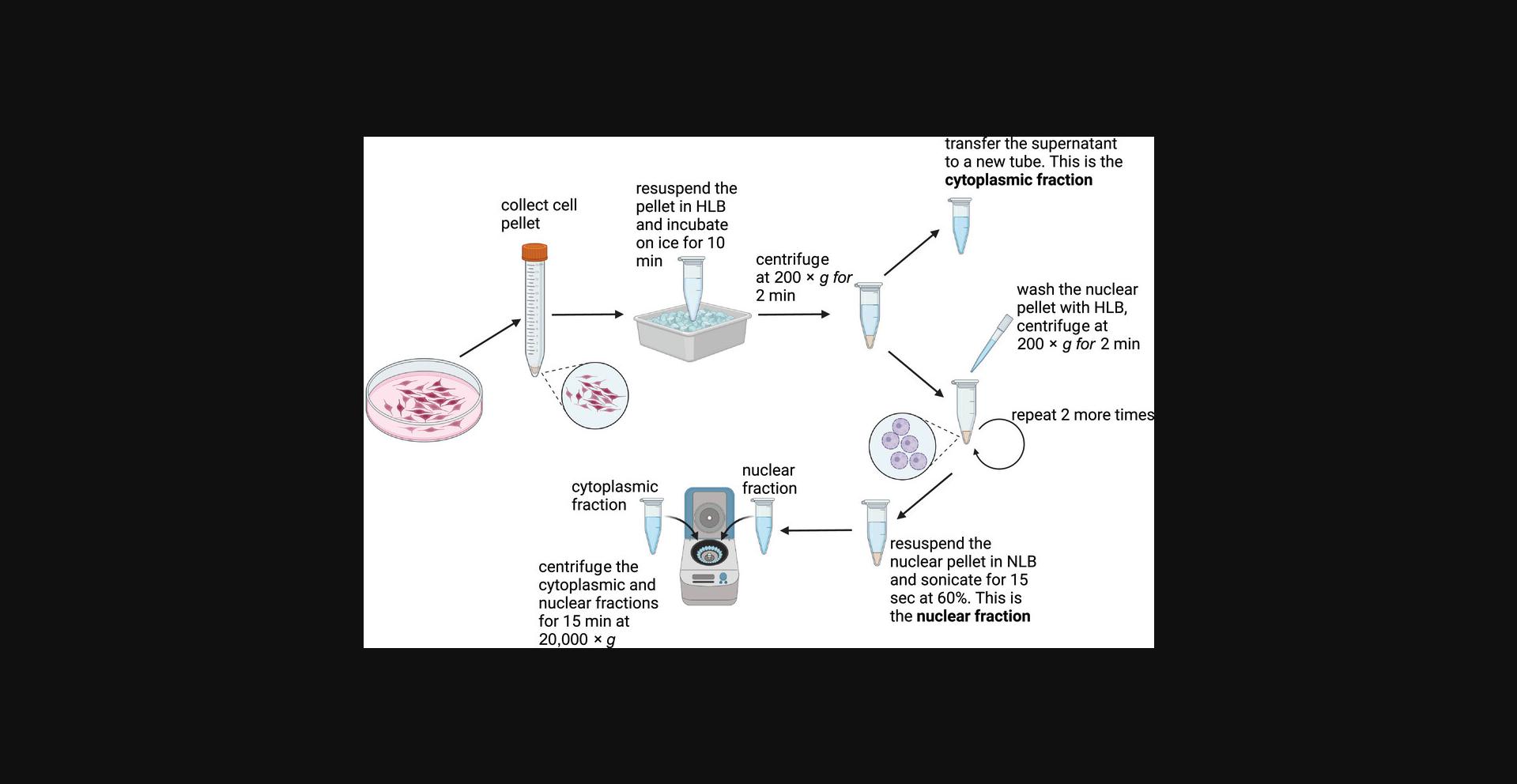
IMPORTANT NOTES : All centrifugation steps should be performed at 4°C. During fractionation, samples must be kept on ice. Resuspension steps must be performed by gentle pipetting to avoid lysing the cells completely.
Materials
-
Mammalian adherent cells, grown to ∼80% confluence on a 10-cm culture dish.
-
Gibco™ Trypsin-EDTA (0.25%), phenol red (Fisher, cat. no.: 11560626)
-
Gibco™ Phosphate-buffered saline (PBS) 1×, pH 7.4 (Fisher, cat. no.: 15374875)
-
HLB buffer (see recipe)
-
NLB buffer (see recipe)
-
Thermo Scientific™ Protease and Phosphatase Inhibitor Cocktail, EDTA-free (100×) (Thermo Scientific™ cat. no.: 78445)
-
Tissue culture dish, 100 × 20 mm (Sarstedt, cat. no.: 83.3902)
-
Corning™ Cell Lifter (Fisher, cat. no.: 10508292)
-
1.5-ml Eppendorf tubes (Fisherbrand, cat. no.: 15432545)
-
Centrifuge: Eppendorf® Centrifuge 5425 R (Merck KGa; cat. no.: EP5406000445) capable of 200 and 21,000 × g
-
Vortex mixer
-
Sonicator: Sonics VCX130 (Merck KGa; cat. no.: Z412619)
Sample preparation
1.Harvest cells from a 10-cm plate by scraping or trypsinization and transfer the cells into 1.5-ml tube.
2.Collect the cell pellet by centrifuging for 5 min at 300 × g.
3.Carefully pipette and discard the supernatant.
4.Wash the cell pellet by adding 500 µl of 1 × PBS followed by centrifuging for 5 min at 300 × g.
5.Carefully pipette and discard the supernatant; do not leave any remaining PBS.
Isolation of cytoplasmic fraction
6.Resuspend the cell pellet in 300 µl HLB buffer by gently pipetting up and down 2-3 times or until the pellet is completely dissolved.
7.Incubate the sample on ice for 10 min.
8.Centrifuge the cell suspension for 2 min at 200 × g.
9.Collect the supernatant as the cytoplasmic fraction into a clean pre-chilled 1.5-ml tube and keep it on ice.
Isolation of nuclear fraction
10.Wash the remaining nuclear pellet with 300 µl of HLB buffer. Gently resuspend the pellet and immediately pellet the cell suspension by centrifuging for 2 min at 200 × g.
11.Pipette the supernatant and discard it; do not leave any remaining buffer.
12.Repeat wash steps 10 and 11 two more times.
13.Resuspend the nuclear pellet in 300 µl NLB buffer.
14.Vortex briefly and then sonicate the sample for 15 s at 60% amplitude.
Collection of cellular fractions and downstream analysis
15.Centrifuge the cytoplasmic fraction (from step 9) and nuclear fraction (from step 14) for 15 min at 20,000 × g.
16.Collect the supernatant into new fresh and pre-chill 1.5-ml tubes. Proceed directly to downstream applications or store sample at −20°C short-term or at −80°C long-time.
17.Measure the protein concentration with Bradford assay following Support Protocol 1.
18.Estimate the quality of fractionation by Western blotting analysis following Support Protocol 2.
Sample Data
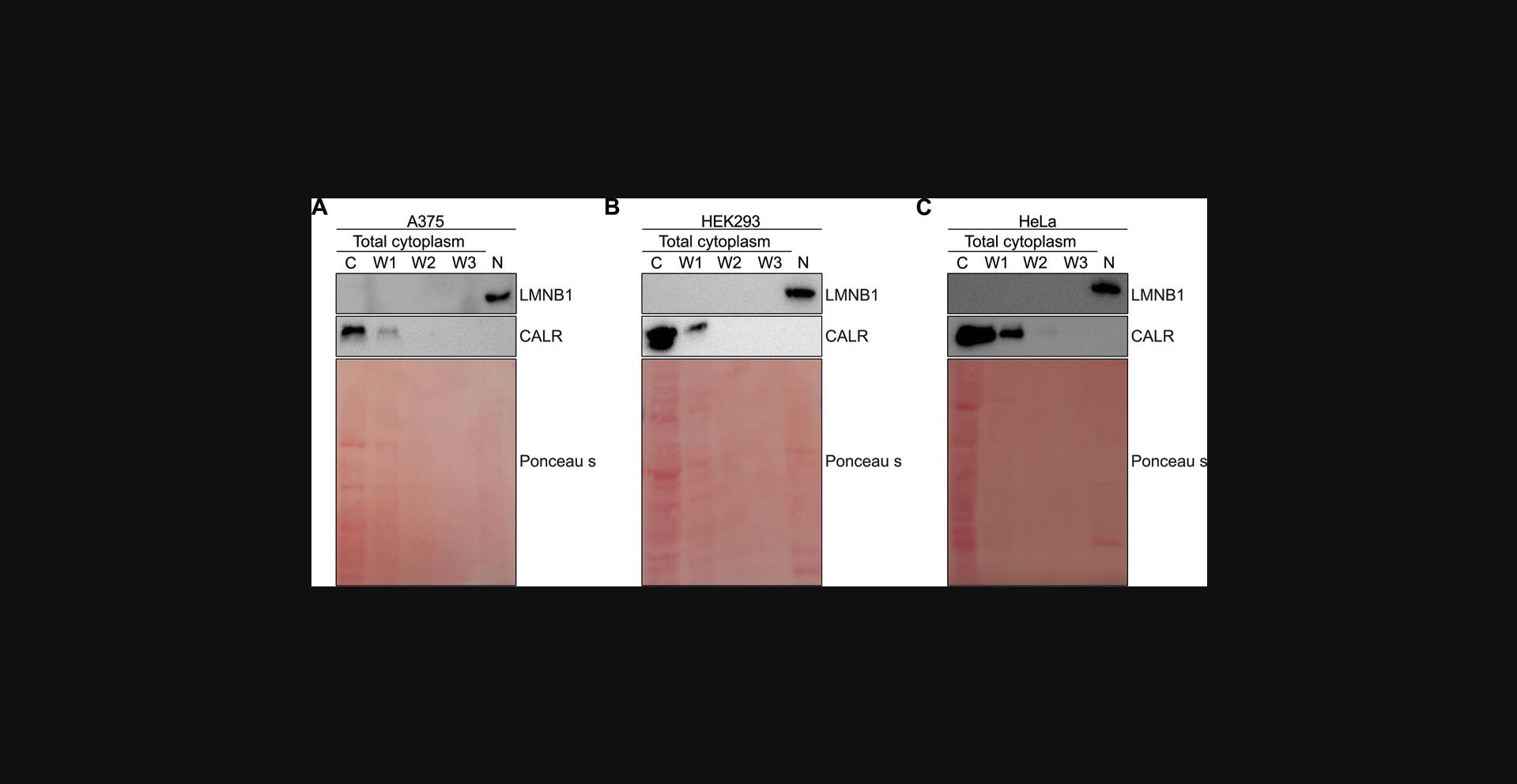
Here we performed subcellular fractionation for A375, HEK293 and HeLa cell lines following the protocol above. To analyze the efficiency of fractionation we investigate the localization of cytoplasmic marker Calreticulin (CALR) and nuclear marker Lamin B1 (LMNB1). During the fractionation, all three washes were collected to demonstrate the effectiveness of the protocol, and the absence of fraction loss during washing steps. We performed Western blot analysis for cytoplasmic and nuclear protein lysates, as well as for all three washes (detailed protocol is described in Support Protocol 2) and probed membranes with CALR (cytoplasmic marker) and LMNB1 (nuclear marker). Our obtained data show the successful fractionation (Fig. 2): the nuclear marker Lamin B1 is presented only in the nuclear fraction in all studied cell lines, while cytoplasmic Calreticulin is predominantly in cytoplasm with minor loss during the first wash.
Support Protocol 1: PROTEIN QUANTIFICATION USING BRADFORD ASSAY
Bradford assay is a method used for measuring the concentrations of proteins (Kielkopf et al., 2020). The Bradford reagent, acidified Coomassie Brilliant Blue G-250, binds to protein and its absorbance maximum moves from 470 to 595 nm (Gallagher & Desjardins, 2011; Kielkopf et al., 2020). When using Bradford assay, protein samples should be diluted in the same buffer they are dissolved in. Herein, we use HLB buffer.
Materials
-
Bovine serum albumin (BSA; Fisher, cat. no.: BP9703)
-
MilliQ water
-
HLB buffer (see recipe)
-
Bradford Reagent (Sigma, cat. no.: B6916)
-
1.5-ml Eppendorf tubes (Fisherbrand, cat. no.: 15432545)
-
Micro test plate, 96-well, transparent (Sarstedt, cat. no.: 82.1582)
-
Microplate reader CLARIOstar (BMG Labtech)
Preparation of standards
1.Prepare 5 mg/ml stock of BSA in MilliQ water and store it at −20°C.
2.In separate tubes prepare BSA standards in HLB buffer ranging from 0.1-1.6 mg/ml using the BSA stock (5 mg/ml) according to Table 1.
| Vial | Volume of HLB buffer (µl) | Volume and source of BSA (µl) | Final BSA concentration (mg/ml) |
|---|---|---|---|
| A | 340 | 160 of stock | 1.6 |
| B | 250 | 250 of vial A dilution | 0.8 |
| C | 250 | 250 of vial B dilution | 0.4 |
| D | 250 | 250 of vial C dilution | 0.2 |
| E | 250 | 250 of vial D dilution | 0.1 |
| F | 500 | 0 | 0 |
3.Pipette 5 µl of each standard into a separate well of 96-well plate.
Measurement of protein concentration
4.Pipette 1 µl of sample into a separate well.
5.Add 250 µl of the Bradford Reagent into each well and carefully pipette up and down, avoiding the formation of bubbles.
6.Incubate the plate for 10 min in the dark and at room temperature.
7.Measure the absorbance at 595 nm using a microplate reader.
8.Calculate the average absorbance value for each BSA standard and sample.
9.Subtract the average absorbance measurement of the blank standard F from the measurements of all other average standards and average sample.
10.Make a standard curve by plotting the average ‘blank–corrected measurement’ for each BSA standard (A – F) versus its concentration in mg/ml.
11.Determine the protein concentration of the sample using the equation for the linearized standard curve.
Sample Data
Here we performed an estimation of protein concentration for cytoplasmic fraction using Bradford assay. The absorbance measurements for BSA standards A – F with downstream calculations are shown in Table 2.The standard curve was plotted using average minus blank values for each BSA standard (A – F) versus its concentration in mg/ml (Fig. 3). The curve was linearized, and the values of sample concentration were obtained by using straight line equation.
| Standard | A | B | C | D | E | F |
|---|---|---|---|---|---|---|
| replicate 1 | 0.804 | 0.639 | 0.502 | 0.438 | 0.409 | 0.378 |
| replicate 2 | 0.907 | 0.627 | 0.494 | 0.435 | 0.412 | 0.375 |
| replicate 3 | 0.866 | 0.646 | 0.551 | 0.463 | 0.412 | 0.388 |
| Average | 0.859 | 0.637 | 0.516 | 0.445 | 0.411 | 0.380 |
| Minus Blank | 0.479 | 0.257 | 0.135 | 0.065 | 0.031 | 0.000 |
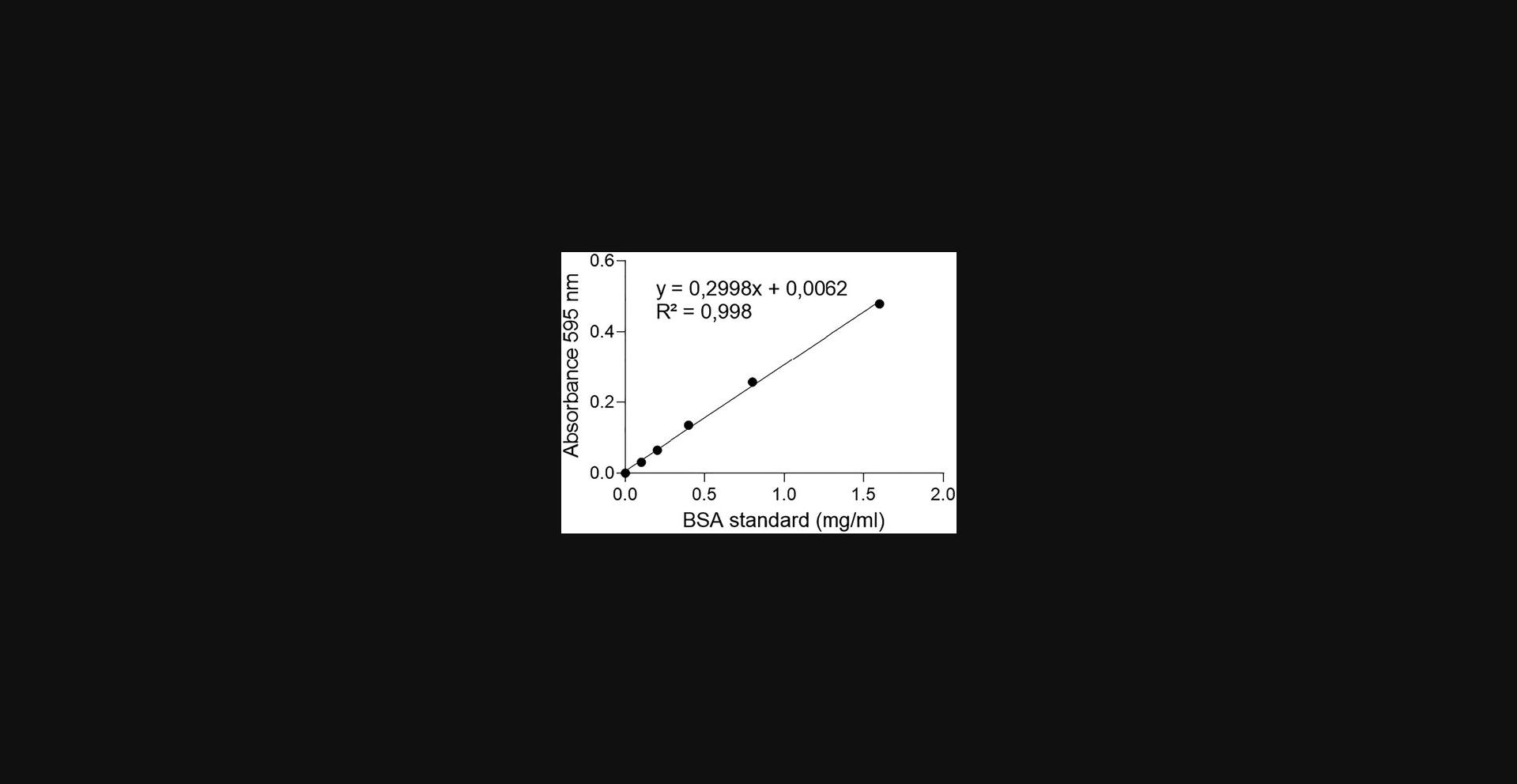
Sample concentration (mg/ml) = (Average Sample absorbance – 0.0062)/0.2998*5
5 is a dilution factor.
Support Protocol 2: SDS-PAGE AND WESTERN BLOTTING
Western blot is a method used to identify proteins from complex lysates. Western blotting separates proteins by molecular weight and in combination with antibodies, target proteins are marked for identification (Mahmood & Yang, 2012; Ni et al., 2016). Here, we describe Western blot analysis for the estimation of the fractionation efficacy. The protocol describes the preparation of homemade SDS-PAGE gel, gel electrophoresis, proteins transfer, probing with antibodies and protein visualization.
Materials
-
Resolving gel (see recipe)
-
Stacking gel (see recipe)
-
Running buffer (see recipe)
-
Laemmli SDS sample buffer, reducing (6×) (Thermo Fisher, cat. no.: J61337.AC)
-
Thermo Scientific™ PageRuler™ Plus Prestained Protein Ladder, 10 to 250 kDa (Thermo Scientific, cat. no.: 11832124)
-
Ponceau S (Thermo Scientific, cat. no.: 6226-79-5)
-
1× TBST (see recipe)
-
2% skim milk (see recipe)
-
Calregulin Antibody (Santa Cruz; cat. no.: sc-373863)
-
Lamin B1 Antibody (Abcam; cat. no.: ab16048)
-
Mouse IgG HRP Linked Whole Ab (Merck KGa; cat. no.: GENA931-1ML)
-
Rabbit IgG HRP Linked Whole Ab (Merck KGa; cat. no.: GENA934-1ML)
-
Supersignal west dura extend duration substrate (Thermo Fisher, cat. no.: 34076)
-
Invitrogen SureCast™ Glass Plates (Invitrogen, cat. no.: HC1001)
-
Invitrogen SureCast™ Gel Handcast Station (Invitrogen, cat. no.: HC1000)
-
15-ml falcon tube (Sarstedt, cat. no.: 62.554.502)
-
1.5 ml Eppendorf tubes (Fisherbrand, cat. no.: 15432545)
-
Gel electrophoresis power supply Fisherbrand Power Pro (Thermo Fisher Scientific; cat. no.: 15818481)
-
Vortex mixer
-
Centrifuge
-
Invitrogen™ Mini Gel Tank and Blot Module Set (Thermo Fisher Scientific; cat. no.: 15384604)
-
Amersham protran premium NC nitrocellulose membranes (Cytiva, cat. no.: 10600003)
-
Thermo Scientific™ Western Blotting Filter Paper, Extra Thick, 20 cm × 20 cm (Thermo Fisher Scientific; cat. no.: 10675935)
-
Semi-dry transfer unit TE70X (Hoefer Inc.)
-
ChemiDoc MP imaging system (Bio-Rad Laboratories, Inc.; cat. no.: 12003154)
1.Use Invitrogen SureCast gel system and assemble glass cassette sandwich for SDS-PAGE gel preparation.
2.Prepare 8% resolving gel fresh in a 15-ml falcon tube and immediately caste the gel.
3.After the polymerization of the resolving gel, prepare the stacking gel, remove the top layer of water, and pour the stacking gel onto the cassette on top of the resolving gel. Quickly add a comb and once the gel has polymerized, proceed to the next step.
4.Place the gel in the gel electrophoreses tank and fill the tank with running buffer.
5.Prepare the protein samples extracted from the cytoplasmic and nuclear fractions by mixing the protein lysate with 6× Laemmli buffer.
6.Denature the sample by heating for 5 min at 95°C.
7.Cool the samples to room temperature, vortex briefly, and collect by centrifugation.
8.Load 2.5 µl of protein ladder and 5-20 µg of cytoplasmic fraction. Load equal volume of nuclear fraction as the cytoplasmic fraction into the wells to maintain a 1:1 ratio.
9.Run the electrophoresis at 70 V while the samples are in the stacking gel. After the samples enter the resolving gel, increase the voltage up to 120 V.
10.Transfer the proteins onto a nitrocellulose membrane using a semi-dry transfer system. The transfer sandwich should be built: (Bottom) Filter paper → Nitrocellulose membranes → Gel → Filter paper (Top)
11.Transfer at 400 mA for 40 min.
12.Once the gel is transferred onto the nitrocellulose membrane, the efficacy of the transfer can be checked by Ponceau S staining. Dip the nitrocellulose membrane in Ponceau S for 2 min. Remove the Ponceau S by washing several times in cold water and finally in 1× TBST.
13.Block the membrane in 2% skim milk for 30 min at room temperature.
14.Dilute the primary antibody in 2% skim milk at 1:100- 1:10,000 dilution depending on the antibody efficiency.
15.Incubate the membrane with primary antibodies for 1 hr at room temperature or overnight at 4°C.
16.Wash the membrane 3 times with 1× TBST for 10 min each time.
17.Dilute the secondary antibodies in 2% skim milk.
18.Incubate 30 min to 1 hr at room temperature under gentil agitation to ensure even distribution of the antibody solution.
19.Wash the membrane three times, each time with 1× TBST for 10 min each time.
20.Detect the protein signal by super signal west dura extended duration substrate, using the ChemiDoc Imaging System.
Sample Data
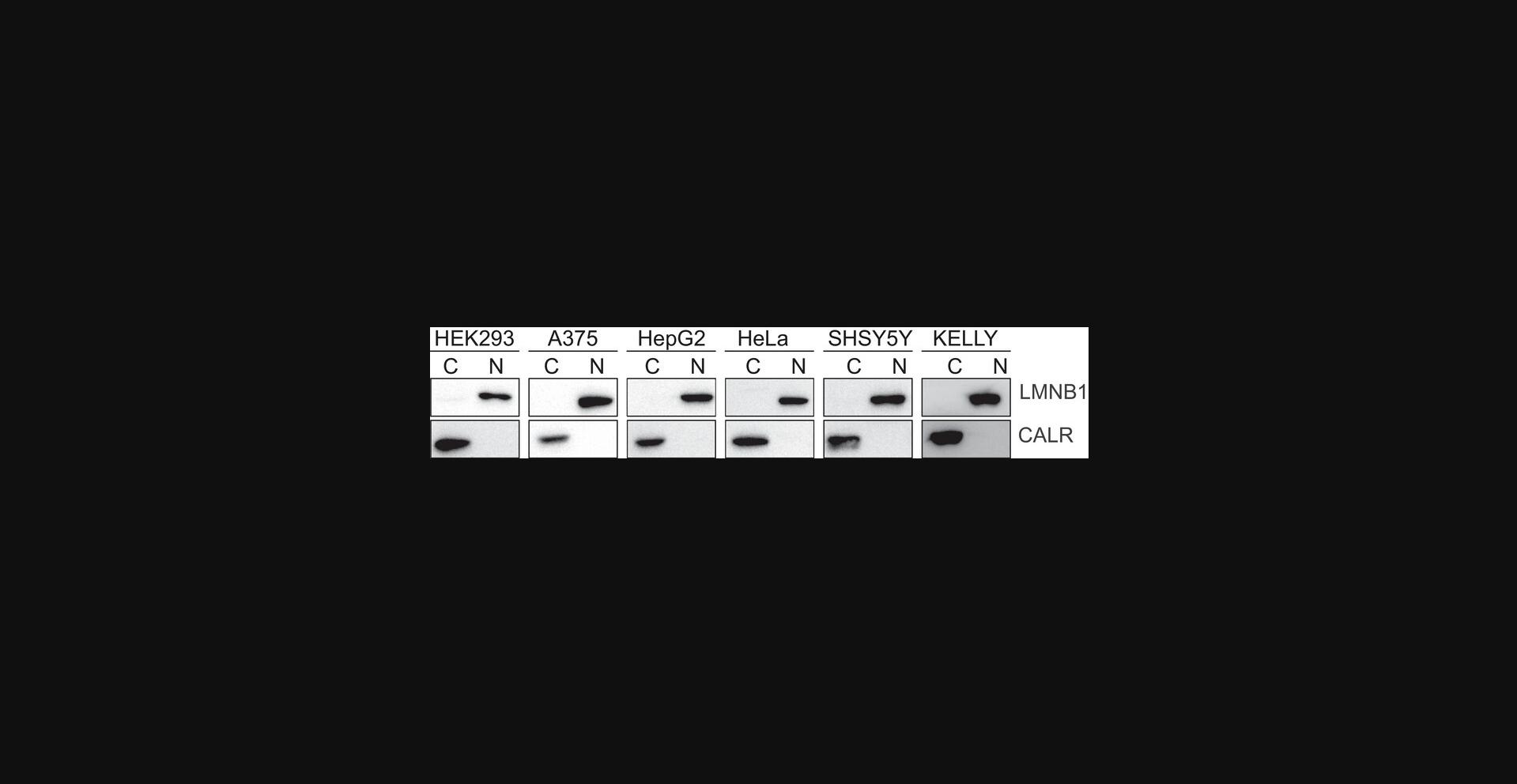
We performed biochemical fractionation of HEK293, A375, HepG2, HeLa, SHSY5Y and KELLY cells using the Basic Protocol (Fig. 4). To evaluate the efficacy of fractionation we performed Western blot analysis of cytoplasmic and nuclear protein lysates. Membranes were probed with Calreticulin and Lamin B1, which showed the effective separation of nuclear and cytoplasmic fractions for most cell lines.
REAGENTS AND SOLUTIONS
Hypotonic lysis buffer (HLB)
| Reagent | Final concentration | Amount (500 ml) |
| 1 M Tris·Cl, pH 7.5 (Invitrogen, cat. no.: 15567) | 10 mM | 5 ml |
|
5 M NaCl (Fisher, cat. no.: BP358) |
10 mM | 1 ml |
|
1 M MgCl2 (Fisher, cat. no.: BP214) |
3 mM | 1.5 ml |
|
NP-40 (Thermo Fisher, cat. no.: J61055) |
0.3% (v/v) | 1.5 ml |
|
Glycerol, 99.5% (Scharlab, cat. no.: GL0026) |
10% (v/v) | 50 ml |
| MQ water | 441 ml | |
| Supplement buffer with 1× proteases and phosphatases inhibitors cocktail (100× stock) | ||
| Store up to 6 months at 4°C | ||
Nuclear lysis buffer (NLB)
| Reagent | Final concentration | Amount (500 ml) |
|
1 M Tris·Cl, pH 7.5 (Invitrogen, cat. no.: 15567) |
20 mM | 10 ml |
|
2 M KCl (Fisher, cat. no.: BP366) |
150 mM | 12.5 ml |
|
1 M MgCl2 (Fisher, cat. no.: BP214) |
3 mM | 1.5 ml |
|
NP-40 (Thermo Fisher, cat. no.: J61055) |
0.3% (v/v) | 1.5 ml |
|
Glycerol, 99.5 % (Scharlab, cat. no.: GL0026) |
10% (v/v) | 50 ml |
| MQ water | 424.5 ml | |
| Supplement buffer with 1× proteases and phosphatases inhibitors cocktail (100× stock) | ||
| Store up to 6 months at 4°C | ||
Resolving gel (8%)
| Total 8 ml (1 gel) | |
|
40% Acrylamide (Invitrogen, cat. no.: HC2040) |
1.6 ml |
|
Resolving buffer (Invitrogen, cat. no.: HC2212) |
2 ml |
| MQ water | 4.3 ml |
|
10% APS (Thermo Fisher, cat. no.: 7727-54-0) |
80 μl |
|
TEMED (Thermo Scientific, cat. no.: 110-18-9) |
8 μl |
| The percentage of the gel can be adjusted by the amount of acrylamide and MQ water | |
| Store up to 6 months at 4°C |
Running buffer
| Reagent | Final concentration | Amount (1000 ml) |
|
250 mM Trizma base (Sigma, cat. no.: 77-86-1) |
25 mM | 100 ml |
| 1.91 M Glycine (Thermo Scientific, cat. no.: 56-40-6) | 191 mM | 100 ml |
|
10% SDS (Millipore, cat. no.: 151-21-3) |
0.1% SDS | 10 ml |
| MQ water | 790 ml | |
| Store up to 6 months at room temperature |
Stacking gel (4%)
| Total 3 ml (1 gel) | |
|
40% Acrylamide (Invitrogen, cat. no.: HC2040) |
0.3 ml |
|
Stacking buffer (Invitrogen, cat. no.: HC2112) |
0.75 ml |
| MQ water | 1.92 ml |
|
10% APS (Thermo Fisher, cat. no.: 7727-54-0) |
30 μl |
|
TEMED (Thermo Scientific, cat. no.: 110-18-9) |
3 μl |
| Store up to 6 months at 4°C |
Skim milk, 2%
| Reagent | Final concentration | Amount (100 ml) |
| Skim milk powder (Millipore, cat. no.: 70166) | 2% (weight/vol) | 2 g |
| MQ water | Up to 1000 ml | |
| Store up to 1 week at 4°C | ||
Transfer buffer
| Reagent | Final concentration | Amount (1000 ml) |
|
250 mM Trizma base (Sigma, cat. no.: 77-86-1) |
25 mM | 100 ml |
|
1.91 M Glycine (Thermo Scientific, cat. no.: 56-40-6) |
191 mM | 100 ml |
|
Methanol (Solveco, cat. no.: 67-56-1) |
20% (v/v) | 200 ml |
| MQ water | 600 ml | |
| Store up to 6 months at room temperature |
TBST, 1×
| Reagent | Final concentration | Amount (1000 ml) |
| TBS tablets (Medicago, cat. no.: 09-7500) | 1× | 2 tablets |
| Tween-20 (Fisher, cat. no.: 9005-64) | 0,1% (v/v) | 1 ml |
| MQ water | Up to 1000 ml | |
| Store up to 6 months at room temperature |
Understanding Results
The purity of the fractionation must be carefully confirmed by western blotting before processing the samples any further. The endoplasmic reticulum protein calreticulin or the house-keeping gene GAPDH can be utilized as a marker for the purification of the cytoplasm, while Histone H3, Lamin A/C, or Lamin B1 can be used as a marker for the nuclear fraction. Consequently, any signal of cytoplasmic marker observed in the nuclear fraction or nuclear marker observed in the cytoplasmic fraction can be considered as contamination.
The main critical point in the processes of biochemical fractionation is to separate the cytoplasmic fraction, while maintaining the nuclei intact. Several washes are needed to clean the nucleus of the remaining cytoplasmic contaminates such as the endoplasmic reticulum, which is attached to the outer nuclear membrane. Greater than 90% of the entire cytoplasm is captured in the first lysis step and very little cytoplasm remains after wash number one (Fig. 2). Gentle pipetting also ensures that the nucleus remains intact and does not leak out to the cytoplasm (Fig. 2). Although it is generally not necessary to keep the washes, it can be recommended in certain cases.
The biochemical procedure outlined herein is carefully adapted to be suitable to a range of cancer cell lines. By optimizing centrifugation parameters, we have obtained a parameter which is broadly applicable and results in high purity and efficiency of fractionation (Fig. 4).
Troubleshooting
See Table 3 for a list of possible problems, causes, and solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| Nuclear marker is detected in the cytoplasmic fraction | Pipetting is too harsh | Gentle pipetting |
| The cytoplasmic fraction is detected in the nuclear fraction | Too big pellet size | Add one more wash step |
| The supernatant looks milky | Pellet does not spin down properly | Split up the fractionation in two different samples |
| The protein concentration is low | Using high volume of the buffer | Adjust the amount of buffer based on the pellet. |
Author Contributions
Hang T. Huynh : Conceptualization; data curation; formal analysis; investigation; methodology; validation; visualization; writing—original draft; writing—review and editing. Evgeniia Shcherbinina : Conceptualization; data curation; formal analysis; investigation; methodology; validation; visualization; writing—original draft; writing—review and editing. Hsiang-Chi Huang : Conceptualization; formal analysis; investigation; methodology; validation; visualization; writing—original draft; writing—review and editing. Reyhaneh Rezaei : Data curation; validation; writing—review and editing. Aishe A. Sarshad : Conceptualization; funding acquisition; investigation; project administration; supervision; writing—original draft; writing—review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Acknowledgments
Work in the Sarshad lab is supported by funding from Knut and Alice Wallenberg Foundation [grant number PAR 2020/228], the Swedish research council [grant number 2019-01855] and the Swedish Society for Medical Research [grant number S19-0019].
Literature Cited
- Alberts, B. (2002). Molecular biology of the cell ( 4th ed.). Garland Science. Publisher description http://www.loc.gov/catdir/enhancements/fy0652/2001054471-d.html
- Fu, X., Liang, C., Li, F., Wang, L., Wu, X., Lu, A., Xiao, G., & Zhang, G. (2018). The rules and functions of nucleocytoplasmic shuttling proteins. International Journal of Molecular Sciences , 19(5), 1445. https://doi.org/10.3390/ijms19051445
- Gagnon, K. T., Li, L., Janowski, B. A., & Corey, D. R. (2014). Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nature Protocols , 9(9), 2045–2060. https://doi.org/10.1038/nprot.2014.135
- Gallagher, S. R., & Desjardins, P. (2011). Quantitation of nucleic acids and proteins. Current Protocols Essential Laboratory Techniques , 5(1), 2.2.1–2.2.36. https://doi.org/10.1002/9780470089941.et0202s5
- Huang, H.-C., Lobo, V., Schön, K., Nowak, I., Westholm, J. O., Fernandez, C., Patel, A. A. H., Wiel, C., Sayin, V. I., Anastasakis, D. G., Angeletti, D., & Sarshad, A. A. (2024). Nuclear RNAi modulates influenza a virus infectivity by downregulating Type-I interferon response. BioRxiv , 2024.2003.2007.583365. https://doi.org/10.1101/2024.03.07.583365
- Jadot, M., Boonen, M., Thirion, J., Wang, N., Xing, J., Zhao, C., Tannous, A., Qian, M., Zheng, H., Everett, J. K., Moore, D. F., Sleat, D. E., & Lobel, P. (2017). Accounting for protein subcellular localization: A compartmental map of the rat liver proteome. Molecular & Cellular Proteomics, 16(2), 194–212. https://doi.org/10.1074/mcp.M116.064527
- Johnson, K. C., Kilikevicius, A., Hofman, C., Hu, J., Liu, Y., Aguilar, S., Graswich, J., Han, Y., Wang, T., Westcott, J. M., Brekken, R. A., Peng, L., Karagkounis, G., & Corey, D. R. (2023). Nuclear localization of Argonaute 2 is affected by cell density and may relieve repression by microRNAs. Nucleic Acids Research , 52, 1930–1952. https://doi.org/10.1093/nar/gkad1155
- Kielkopf, C. L., Bauer, W., & Urbatsch, I. L. (2020). Bradford assay for determining protein concentration. Cold Spring Harbor Protocols , 2020(4), 102269. https://doi.org/10.1101/pdb.prot102269
- Liu, Y., Li, P., Fan, L., & Wu, M. (2018). The nuclear transportation routes of membrane-bound transcription factors. Cell Communication and Signaling , 16(1), 12. https://doi.org/10.1186/s12964-018-0224-3
- Lobo, V., Nowak, I., Fernandez, C., Muler, A. I. C., Westholm, J. O., Huang, H.-C., Fabrik, I., Huynh, H. T., Shcherbinina, E., Poyraz, M., Härtlova, A., Benhalevy, D., Angeletti, D., & Sarshad, A. A. (2024). Loss of Lamin A leads to the nuclear translocation of AGO2 and compromised RNA interference. BioRxiv , 2023.2006.2005.543674. https://doi.org/10.1101/2023.06.05.543674
- Mahmood, T., & Yang, P. C. (2012). Western blot: Technique, theory, and trouble shooting. North American Journal of Medicine and Science , 4(9), 429–434. https://doi.org/10.4103/1947-2714.100998
- Ni, D., Xu, P., Sabanayagam, D., & Gallagher, S. R. (2016). Protein blotting: Immunoblotting. Current Protocols Essential Laboratory Techniques , 12(1), 8.3.1–8.3.40. https://doi.org/10.1002/9780470089941.et0803s12
- Sarshad, A. A., Juan, A. H., Muler, A. I. C., Anastasakis, D. G., Wang, X., Genzor, P., Feng, X., Tsai, P. F., Sun, H. W., Haase, A. D., Sartorelli, V., & Hafner, M. (2018). Argonaute-miRNA complexes silence target mRNAs in the nucleus of mammalian stem cells. Molecular Cell , 71(6), 1040–1050.e1048. https://doi.org/10.1016/j.molcel.2018.07.020
- Senichkin, V. V., Prokhorova, E. A., Zhivotovsky, B., & Kopeina, G. S. (2021). Simple and efficient protocol for subcellular fractionation of normal and apoptotic cells. Cells , 10(4), 852. https://doi.org/10.3390/cells10040852

