Bacterial genome annotation script using BLASTN
Ana Mariya Anhel, Lorea Alejaldre, Ángel Goñi-Moreno
Abstract
This protocol uses a python based script and command-line blastn to annotate Sanger sequencing results from genome amplifications. Its main use in our lab (https://biocomputationlab.com) is to identify the location and gene locus of transposon inserts in microbial bacterial genomes of Pseudomonas putida KT2440. However, this script can be used for other bacterial genomes for which its genome sequence and annotation are available.
Script was developed in python 3.9 with blastn version 2.2.18.
Before start
To run this script command-line blastn and python 3 with packages sys, pandas and os must be installed.
Steps
Annotation of sequencing reads
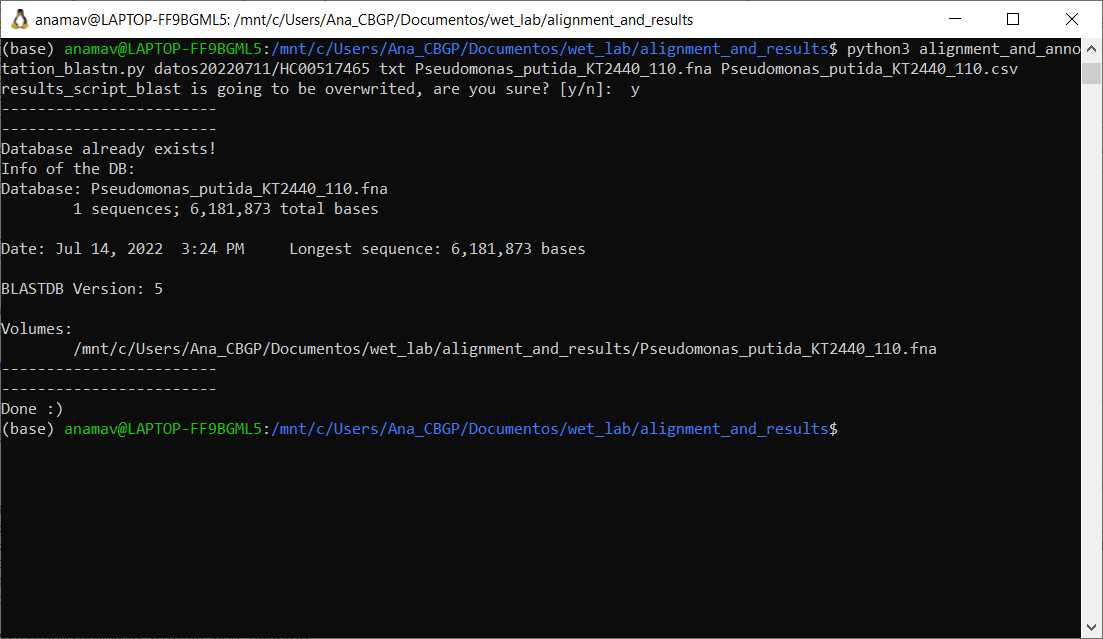
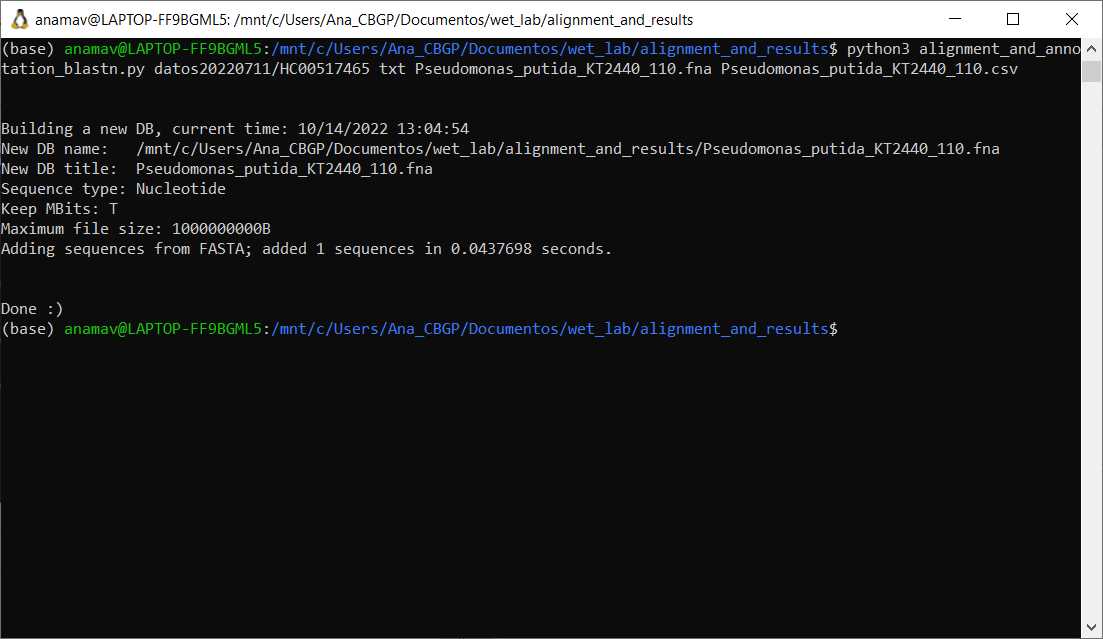
Download genome file in FASTA format and annotation file in .csv for the microbial organism to use as reference
Run the following python based script with the required arguments
#Command to run blastn annotation script
python alignment_and_annotation_blastn.py [directory of sequencing reads] [type of file] [genome file in fasta format] [annotation file in csv format]
Output is a folder named results_script_blast which contains three files:
- all_seq_aligned.sam
- all_seq_aligned.txt
- table_reads_genes_description.csv
Example: Annotation of sequencing results from P. putida KT2440
Input files
- Directory of sequencing reads (it is a zip but shoul be a directory) HC00517465.zip In this case the type of file (extension) is txt
- Genome in FASTA format Pseudomonas_putida_KT2440_110.fna
- Annotation file of that genome Pseudomonas_putida_KT2440_110.csv
Output files
A new folder named results_script_blast (output files attached in the following zip file) contains a table with information about the alignment and genomic context of each sequencing read.
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| query acc. | s. start | % identity | alignment length | mismatches | gap opens | evalue | bit score | subject strand | Locus Tag | Feature Type | Start | End | Strand | Gene Name | Product Name | Subcellular Localization [Confidence Class] | Multiple Allignments | Rest of Locus Tag Associated |
| H220707-054_B23_219DZAA034_premix.ab1 | 6170239 | 99.04299999999999 | 209 | 1 | 1 | 1.8599999999999997e-103 | 374 | plus | PP_5408 | CDS | 61691130 | 61702940 | - | - | hypothetical protein | Cytoplasmic [Class 3] | FALSO | - |
| H220707-054_P21_219DZAA035_premix.ab1 | 6170230 | 98.618 | 217 | 1 | 2 | 2.9799999999999995e-106 | 383 | plus | PP_5408 | CDS | 61691130 | 61702940 | - | - | hypothetical protein | Cytoplasmic [Class 3] | FALSO | - |
| H220707-054_L21_219DZAA036_premix.ab1 | 6170230 | 99.539 | 217 | 0 | 1 | 1.4899999999999998e-109 | 394 | plus | PP_5408 | CDS | 61691130 | 61702940 | - | - | hypothetical protein | Cytoplasmic [Class 3] | FALSO | - |
| H220707-054_F19_219DZAA037_premix.ab1 | 6170230 | 99.083 | 218 | 1 | 1 | 1.9699999999999993e-108 | 390 | plus | PP_5408 | CDS | 61691130 | 61702940 | - | - | hypothetical protein | Cytoplasmic [Class 3] | FALSO | - |
| H220707-054_H21_219DZAA038_premix.ab1 | 6170546 | 97.22200000000001 | 108 | 1 | 2 | 1.3e-45 | 182 | minus | PP_5409 | CDS | 61704660 | 61723010 | - | glmS | L-glutamine/D-fructose-6-phosphate aminotransferase | - | FALSO | - |
| H220707-054_P19_219DZAA039_premix.ab1 | 6170546 | 98.148 | 108 | 0 | 2 | 2.8e-47 | 187 | minus | PP_5409 | CDS | 61704660 | 61723010 | - | glmS | L-glutamine/D-fructose-6-phosphate aminotransferase | - | FALSO | - |
| H220707-054_L19_219DZAA040_premix.ab1 | 6170547 | 96.33 | 109 | 1 | 3 | 6.389999999999999e-44 | 176 | minus | PP_5409 | CDS | 61704660 | 61723010 | - | glmS | L-glutamine/D-fructose-6-phosphate aminotransferase | - | FALSO | - |
| H220707-054_N21_219DZAA041_premix.ab1 | 6170546 | 99.074 | 108 | 0 | 1 | 5.679999999999999e-49 | 193 | minus | PP_5409 | CDS | 61704660 | 61723010 | - | glmS | L-glutamine/D-fructose-6-phosphate aminotransferase | - | FALSO | - |
| H220707-054_J19_219DZAA046_premix.ab1 | 6170533 | 96.84200000000001 | 95 | 0 | 3 | 9.219999999999999e-38 | 156 | minus | PP_5409 | CDS | 61704660 | 61723010 | - | glmS | L-glutamine/D-fructose-6-phosphate aminotransferase | - | FALSO | - |
| H220707-054_D21_219DZAA047_premix.ab1 | 6170239 | 99.51700000000001 | 207 | 1 | 0 | 1.4899999999999996e-104 | 377 | plus | PP_5408 | CDS | 61691130 | 61702940 | - | - | hypothetical protein | Cytoplasmic [Class 3] | FALSO | - |
Final table of the alignment with the correspondant gene or locus insertion