Axon tracing and node segmentation of myelinated bladder afferents in the pelvic nerve
John-Paul Fuller-Jackson, Peregrine B Osborne, Janet R Keast
Abstract
This protocol describes method for tracing myelinated axons labelled with the adeno-associated virus, AAV-PHP.S and neurofascin immunohistochemistry (to identify paranodes) in pelvic nerve of rat, using Neurolucida 360. This tracing method can be applied to various types of peripheral nerves. In rats, AAV-PHP.S has a high tropism for myelinated but not unmyelinated afferents. Cholera toxin subunit B (CTB) microinjected into the bladder was used to identify bladder afferents in the pelvic nerve. This protocol does not include details of the immunohistochemistry or procedures involving animals (viral labelling and CTB tracing). Other antibodies relevant to myelinated axons can be used instead of neurofascin.
Steps
Image acquisition
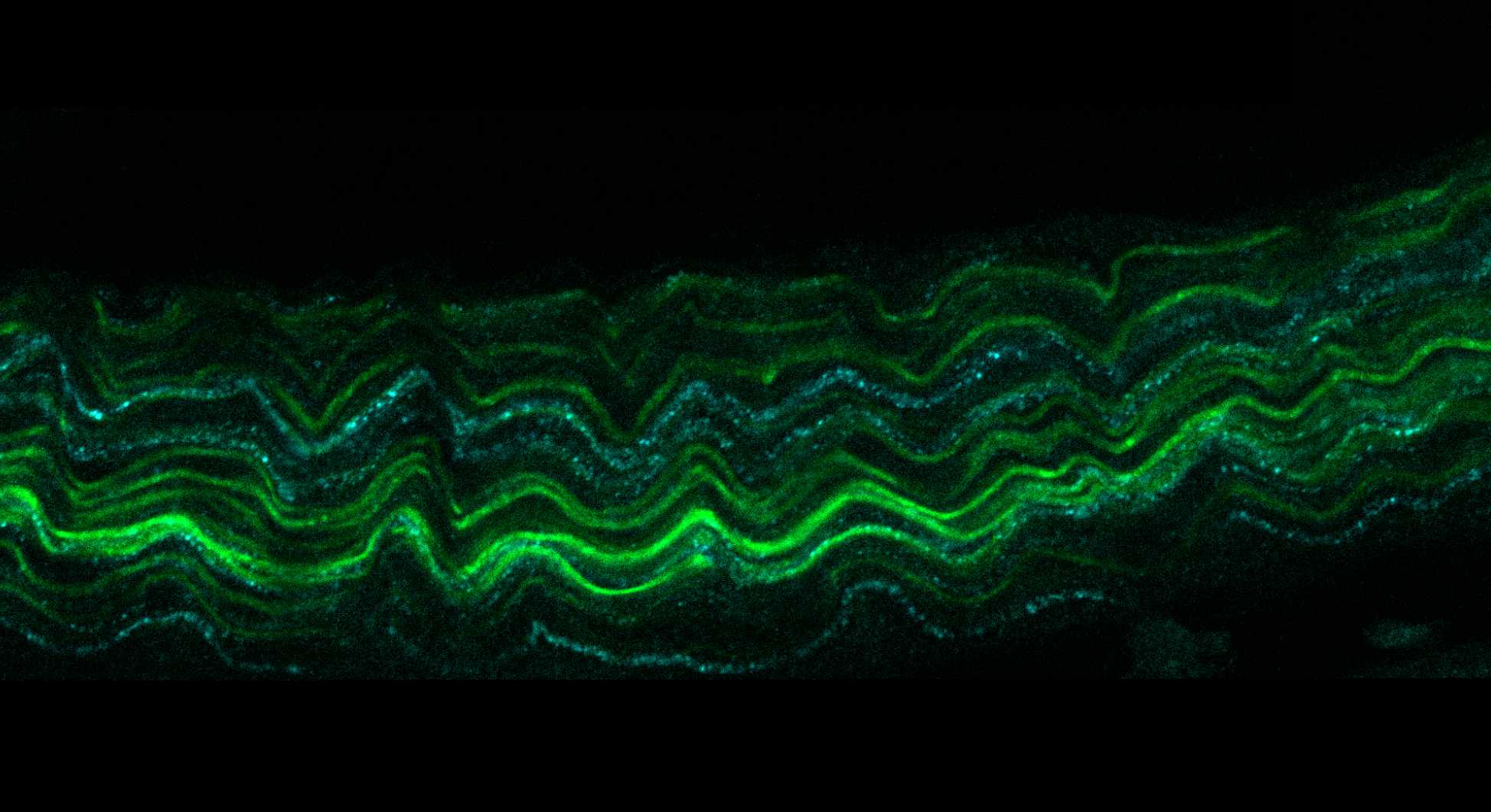
On a confocal microscope, scan the nerve fascicle with sufficient magnification and resolution to achieve a voxel size of at least 0.099 x 0.099 x 1 μm (XYZ). This will require multiple tiles acquired with 10% overlap.
Image pre-processing
Stitch together the tiled image stack dataset into a new single image.
Convert the stitched image to JPEG2000 (JPX) using MicroFile+.
Software
| Value | Label |
|---|---|
| MicroFile+ (RRID: SCR_018724) | NAME |
| MBF Bioscience | DEVELOPER |
Opening file in Neurolucida
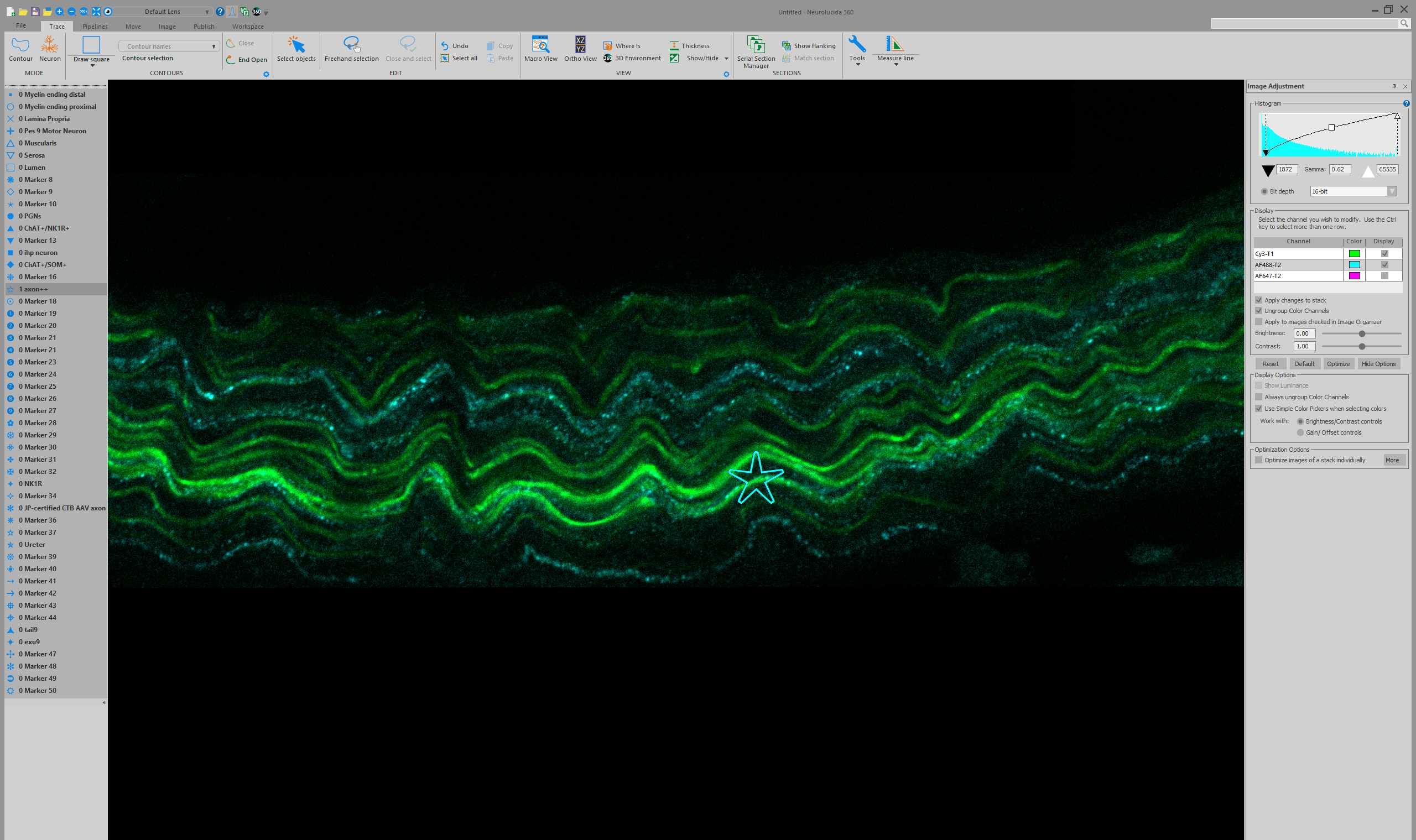
Open the converted nerve JPEG2000 dataset in Neurolucida 360 .
Software
| Value | Label |
|---|---|
| Neurolucida 360 | NAME |
| MBF Bioscience | DEVELOPER |
| https://www.mbfbioscience.com/neurolucida360 | LINK |
| v.2020.1.1 | VERSION |
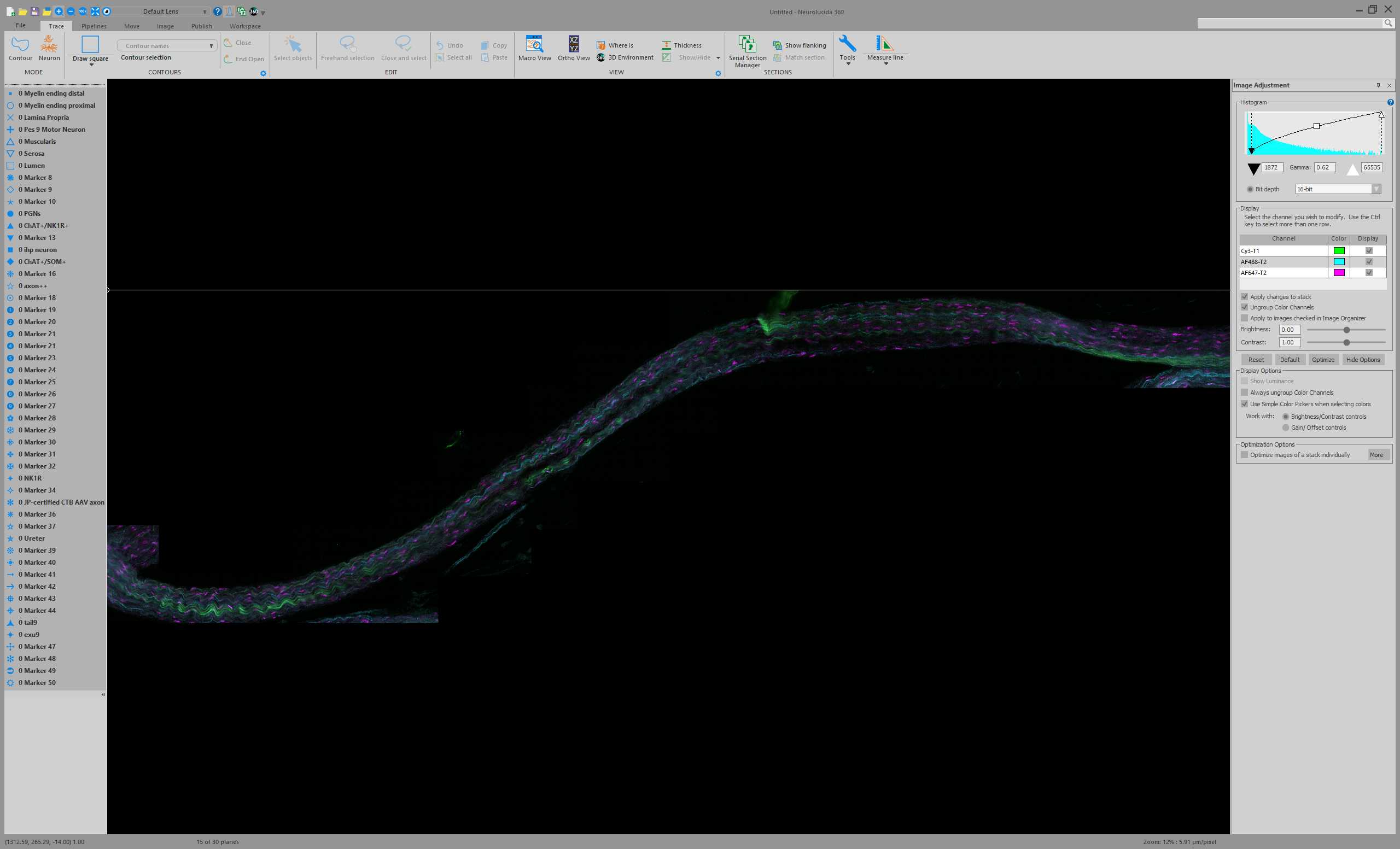
Thoughout all the subsequent Steps, save progress. This will prompt a saving of the JPEG2000 (if display settings were altered) and the saving of a data file. Choose .XML as the file type for saving the data file. Keep the .XML in the same file location as the associated JPEG2000.
Identification of specific axon classes
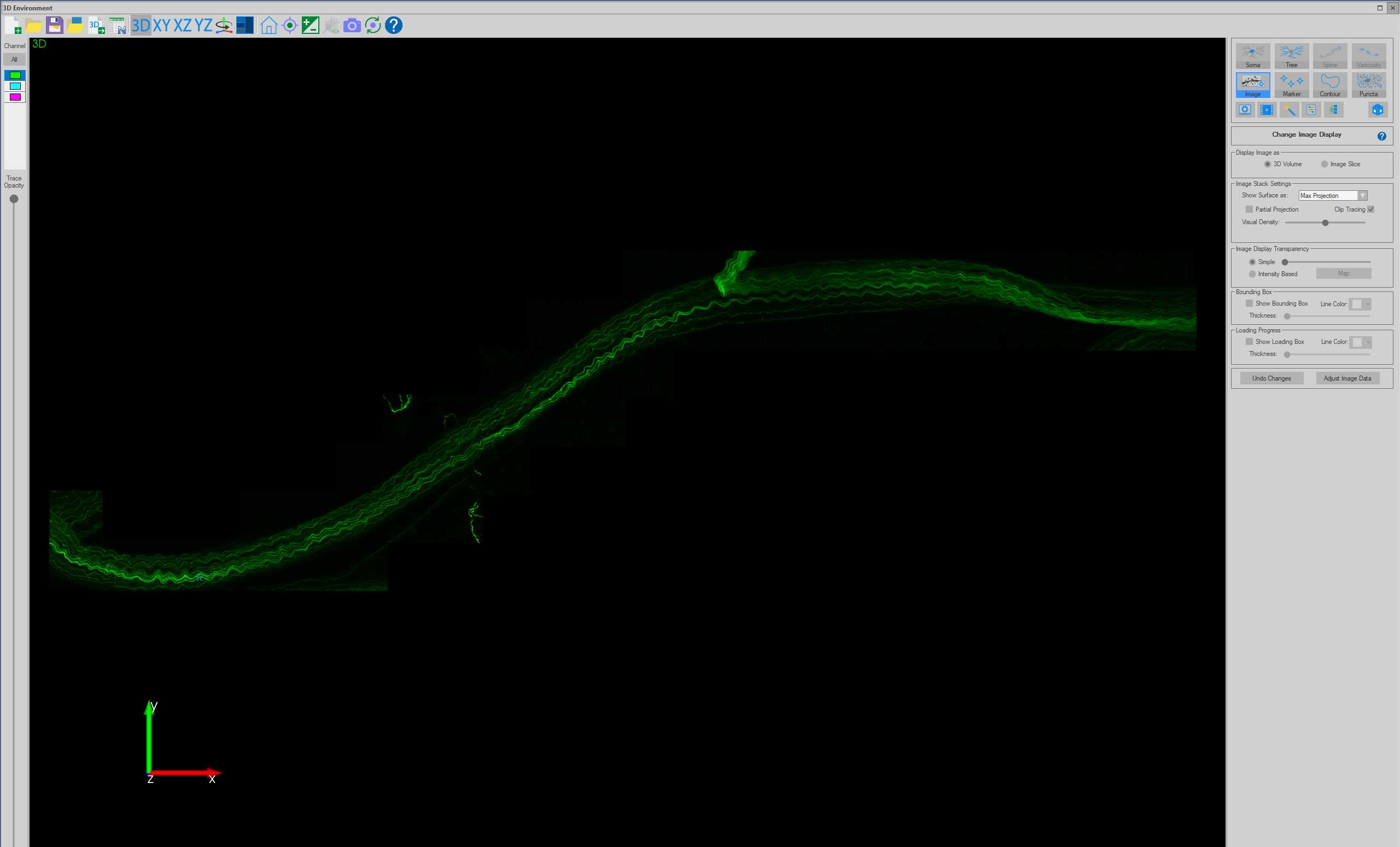
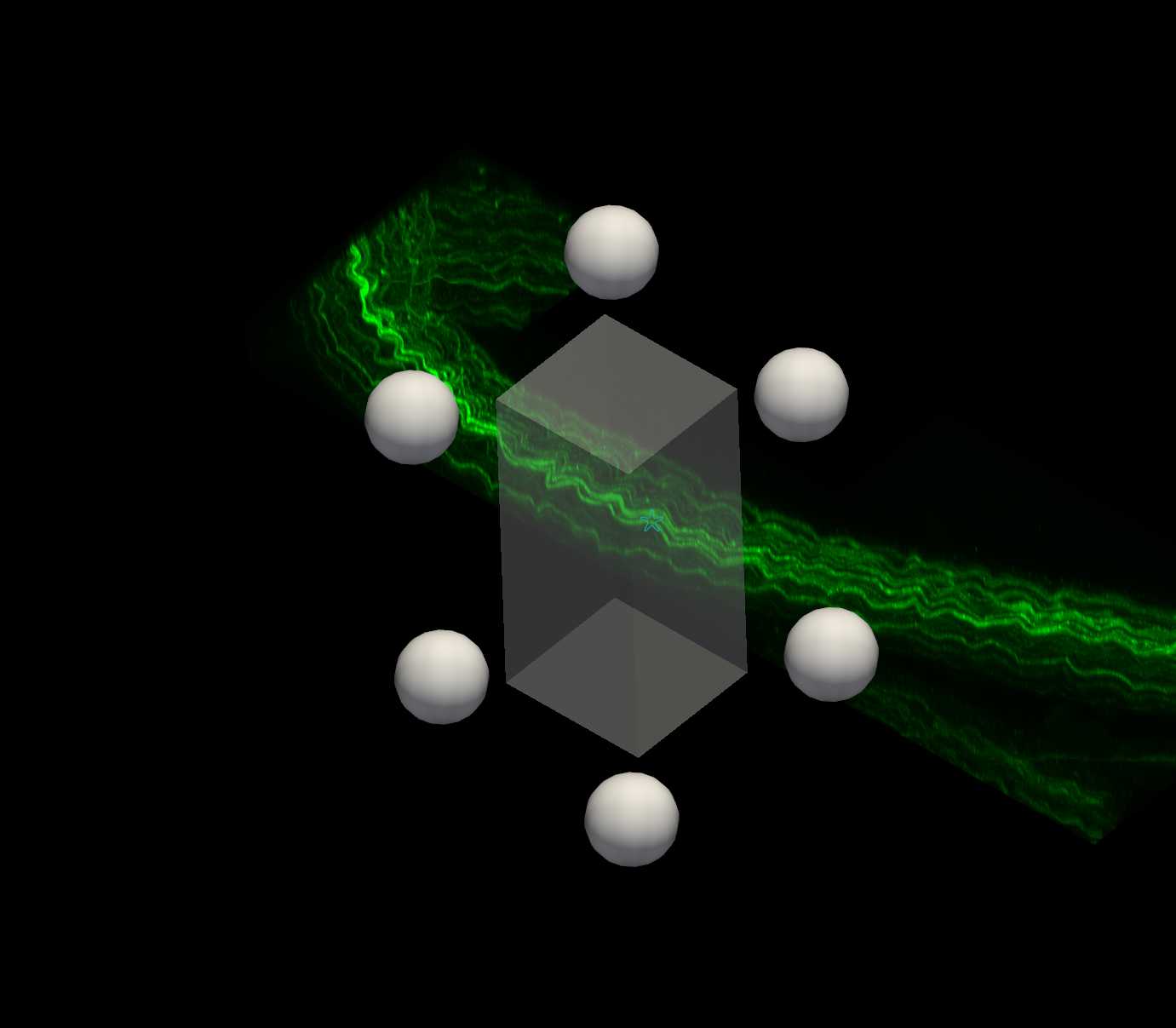
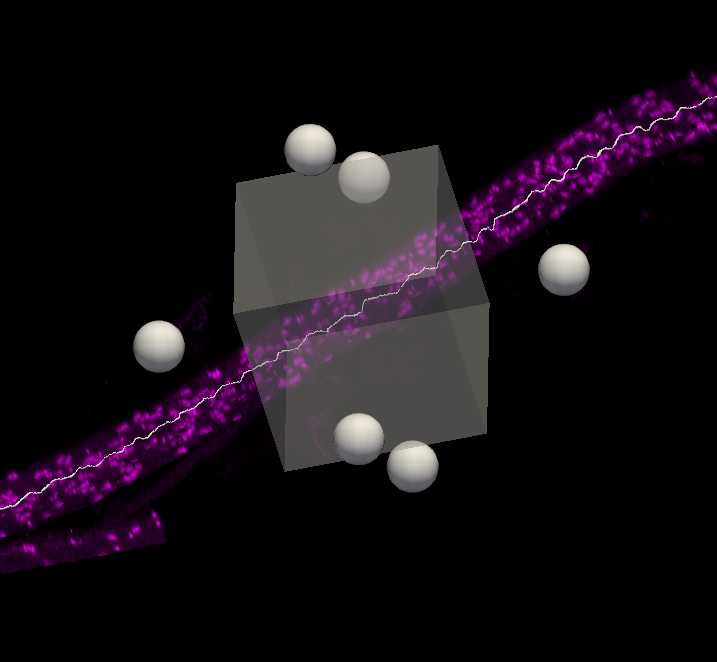
Subvolume 3D viewing of axons
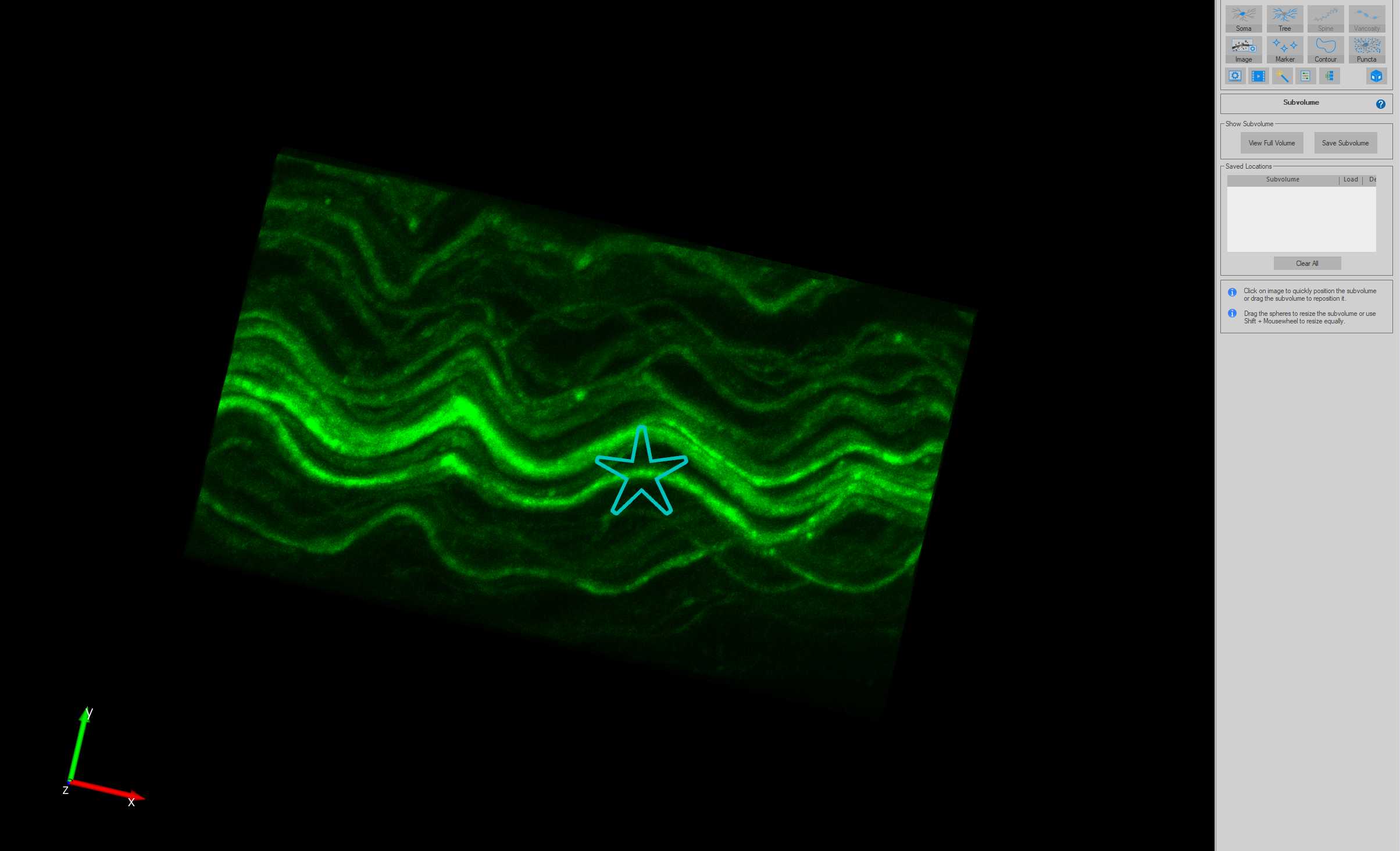
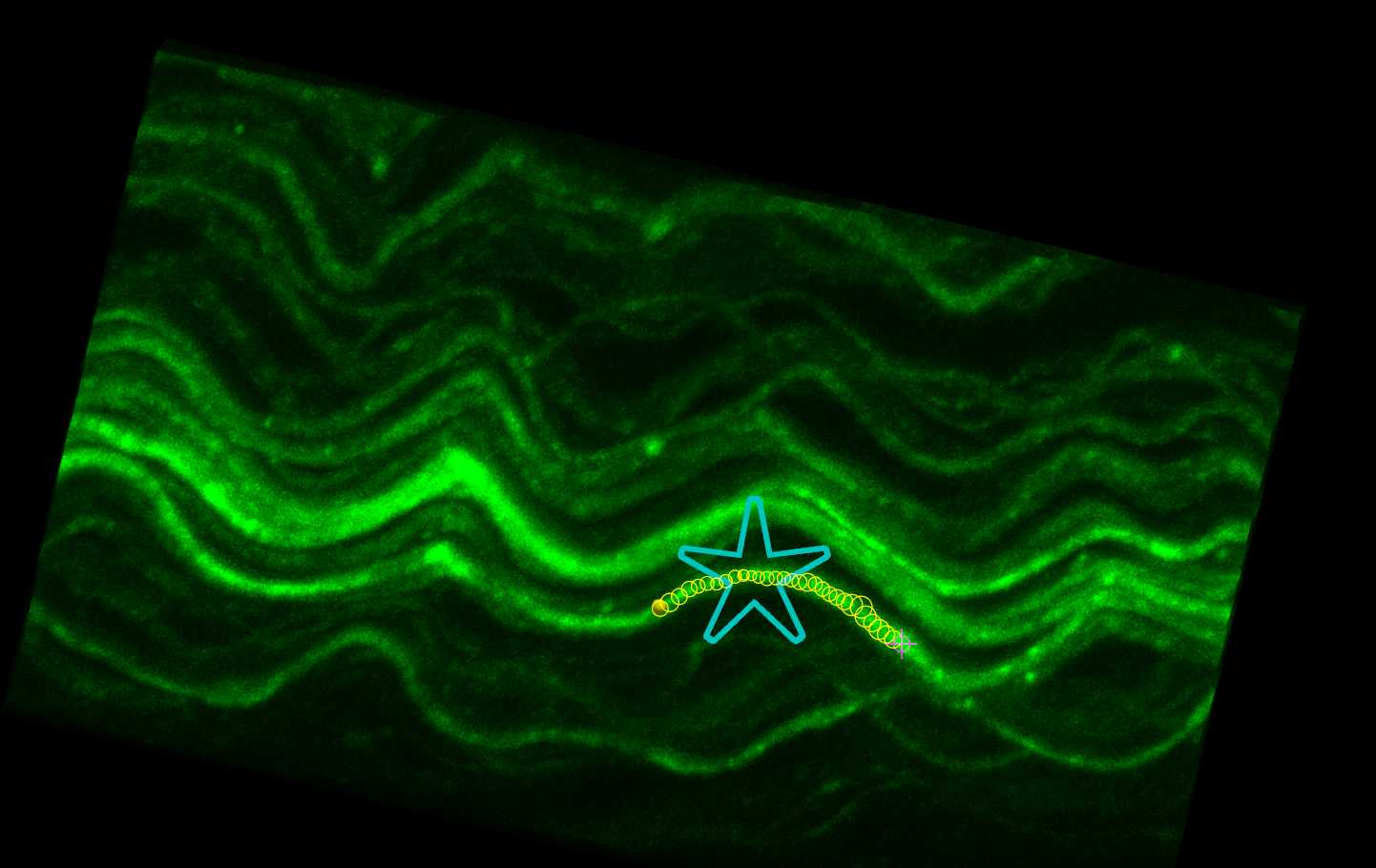
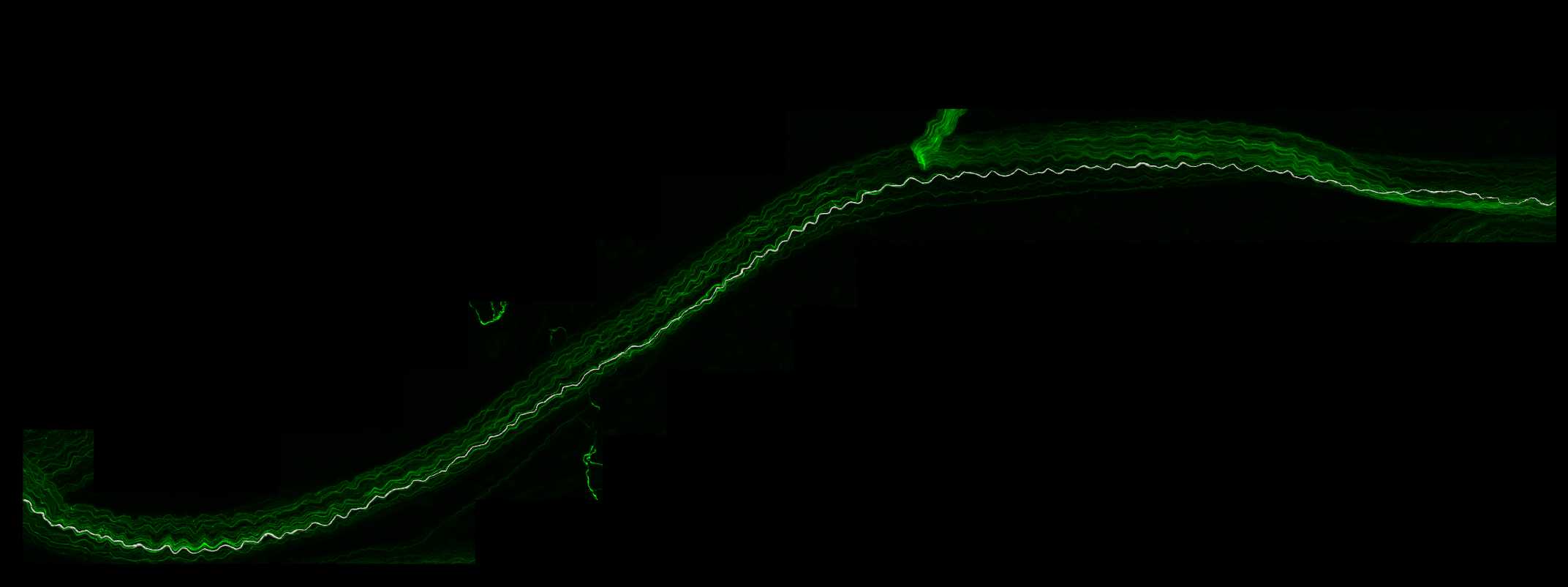
Tracing axons using AAV-PHP.S fluorescence
When the tracing approaches the edge of the Subvolume region, the Subvolume region will automatically shift in the direction of the tracing to allow for seamless tracing of axons without the need to view the entire image stack. Continue tracing until the end of the axon (or when the axon cannot confidently be traced).
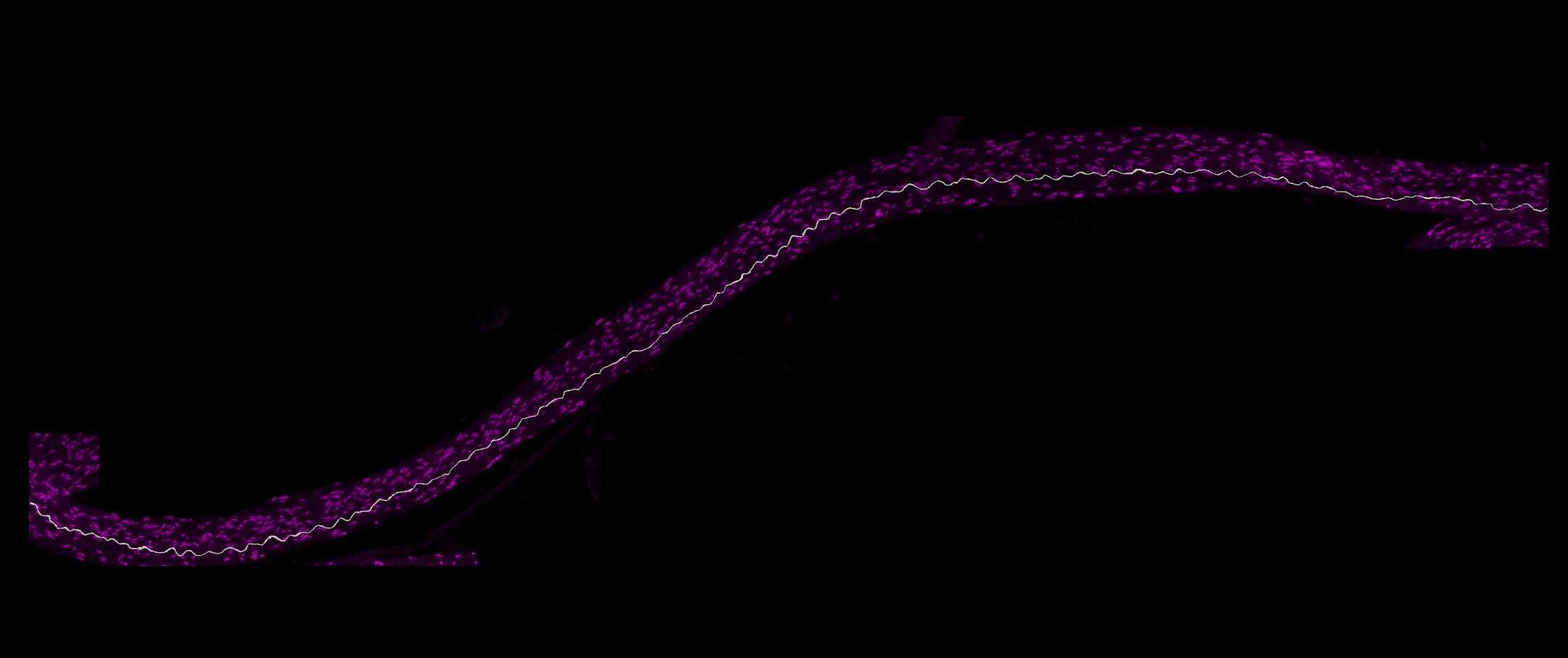
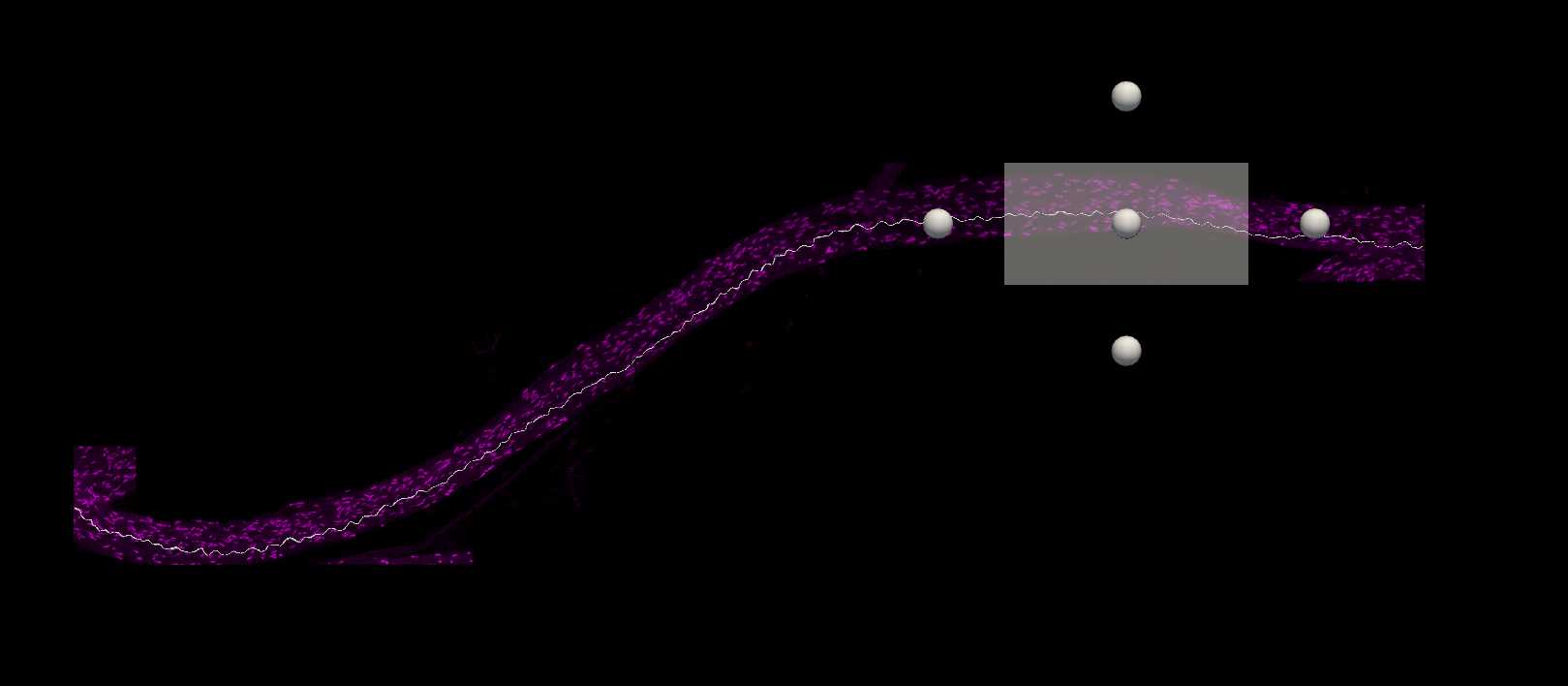
Paranode segmentation on traced axons
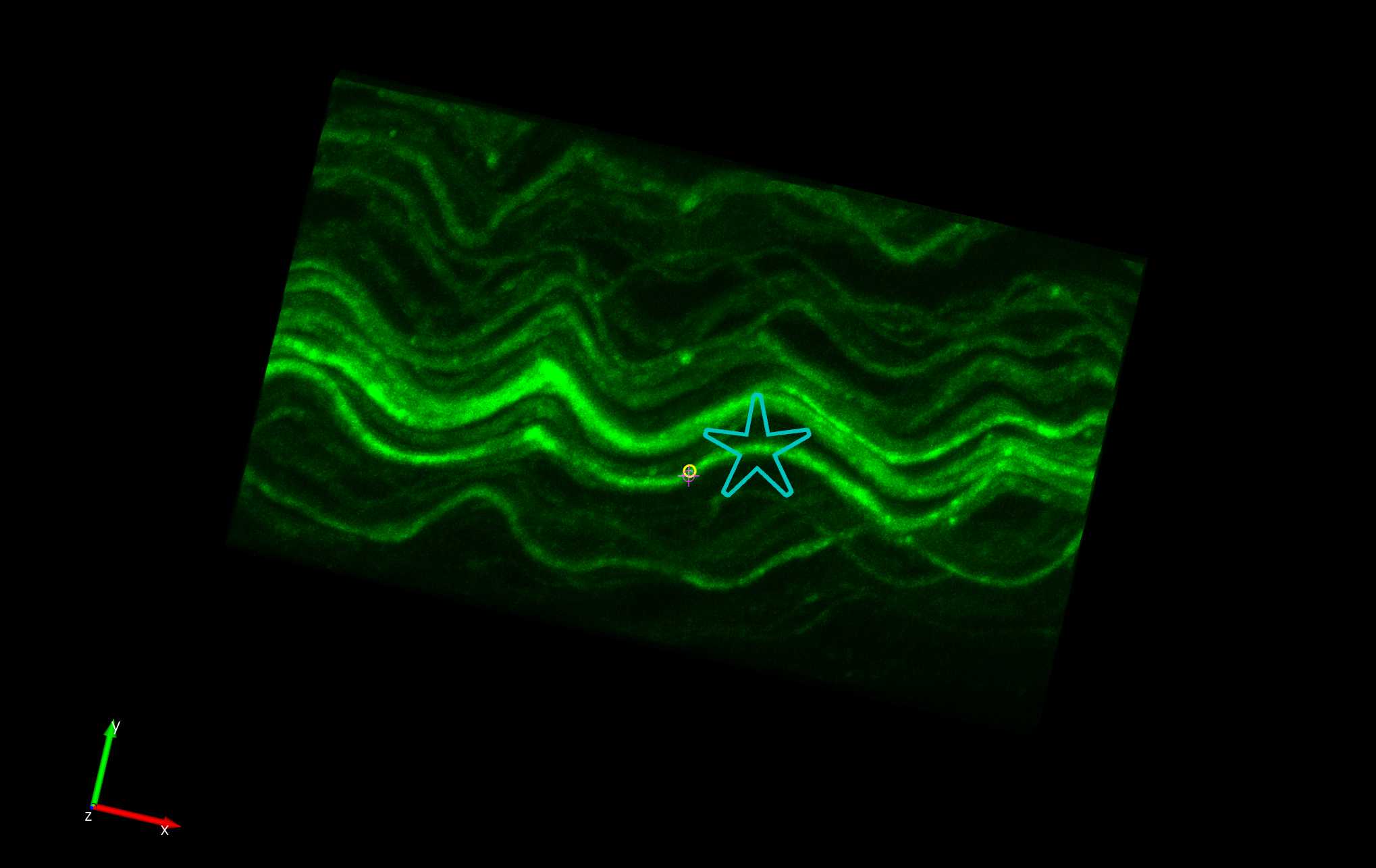
In the Subvolume region, search along the traced axon for Neurofascin labelling that completely overlaps with the axon. Move the image to see the paranodes from multiple angles to ensure correct identification of nodes on the axon. Move the Subvolume region along the axon progressively until a paranode is found.
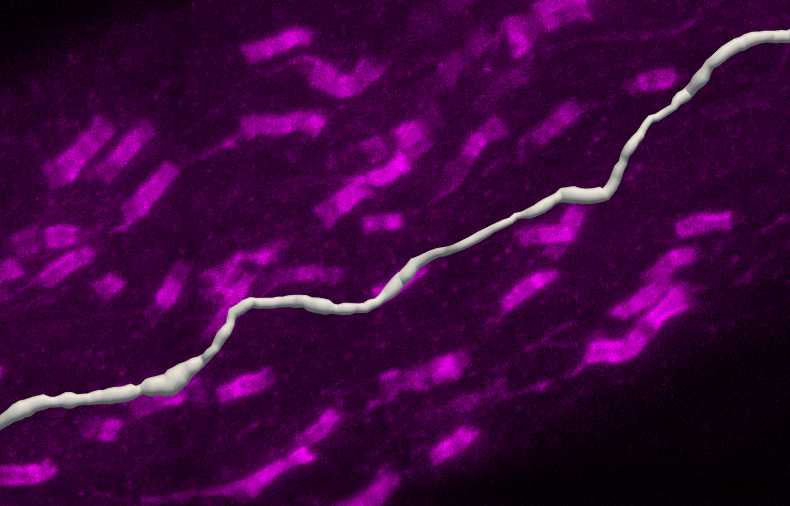
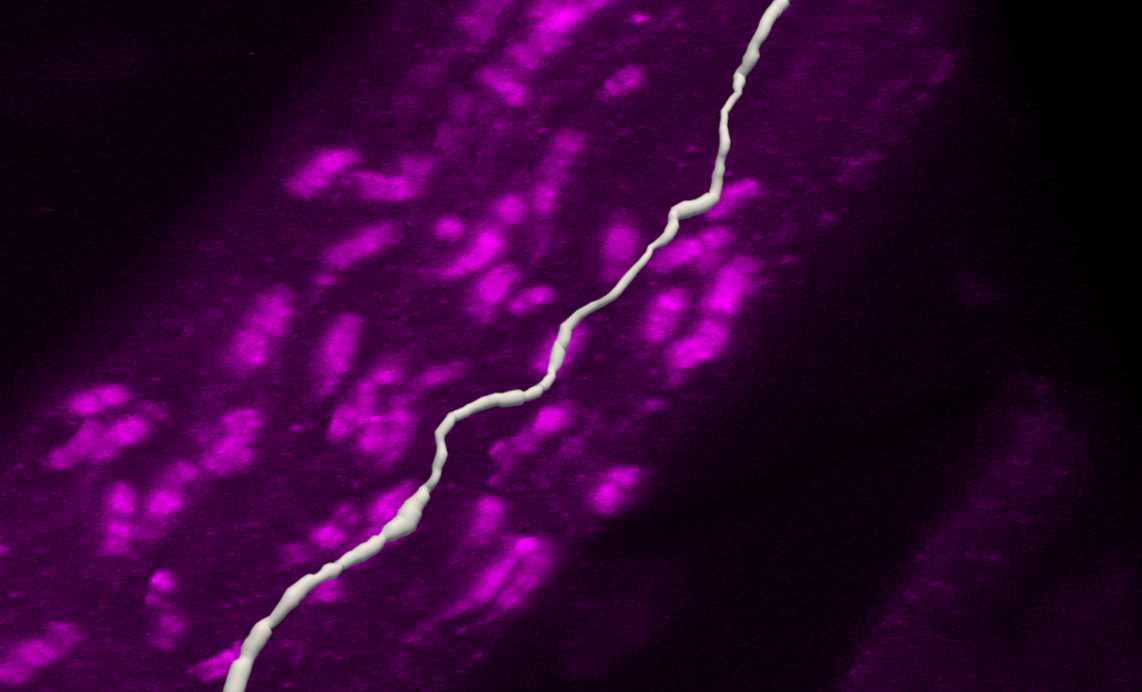
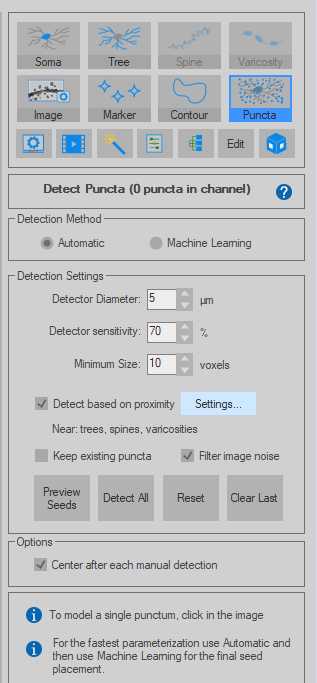
Left click on the paranode in the image to segment that specific paranode. This will create a Punctum that represents the paranode.
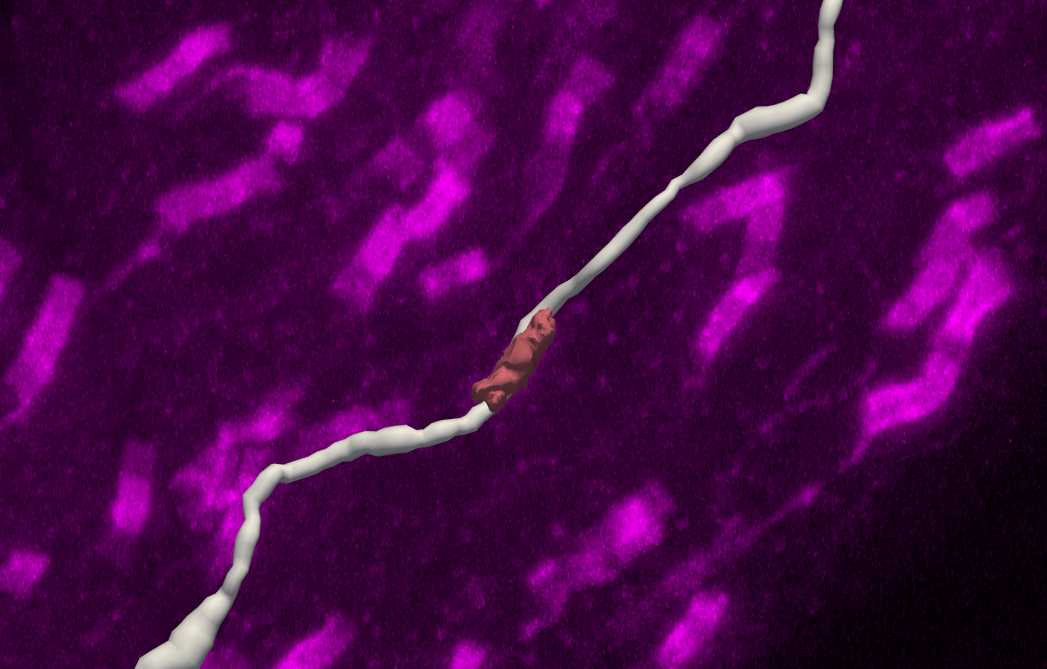
If the detection fails, this is unlikely to be a paranode along the axon (too far away). Sometimes Detector Sensitivity can be adjusted for signal that is not being detected but is clearly along the axon.
Axon and paranode data extraction
To extract data, open the .XML file created by Neurolucida 360 in Neurolucida Explorer .
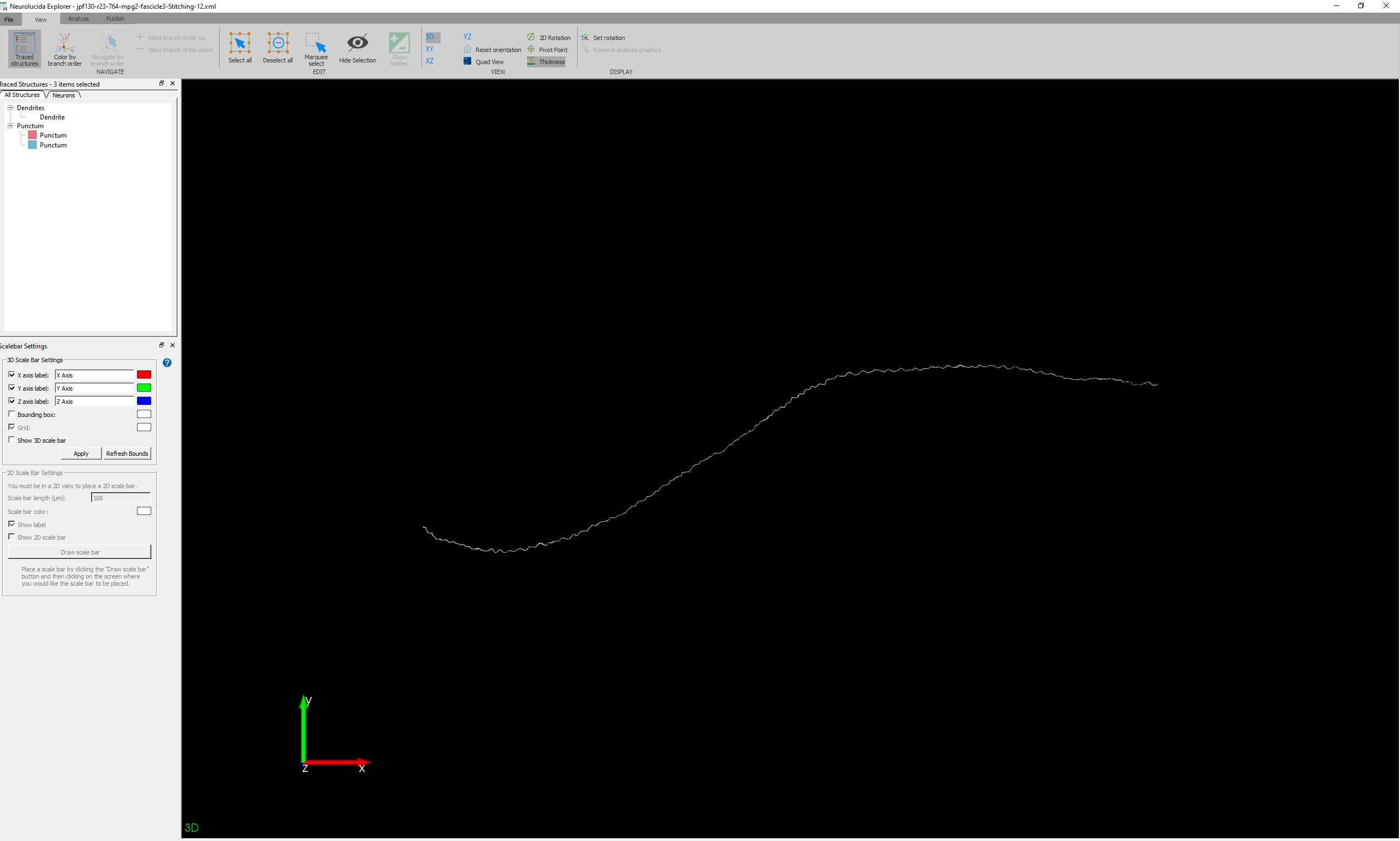
This can be done directly from the 3D Environment of Neurolucida 360 by clicking on the icon that looks like spreadsheet with a superimposed 'N'. This will open the current .XML in Neurolucida Explorer .
Software
| Value | Label |
|---|---|
| Neurolucida Explorer | NAME |
| MicroBrightField Bioscience | DEVELOPER |
| https://www.mbfbioscience.com/neurolucida-explorer | LINK |