Automated QIAcube DNA Extraction
Clemens Scherzer
Abstract
This protocol explains the Standard Operating Protocol for automated extraction of DNA using QIAcube.
Before start
Based on 12 Samples.
DNA Q/C GOALS
- Cary Concentration Assay
1. 260/280 = 1.8-2.0
2. Manual Puragene Extraction: 260 µg /mL (65 µg total) of DNA/subject
3. Automated QIAcube Extraction: 125 µg/mL (50 µg total) of DNA/subject
2. .7% Agarose Gel Electrophoresis
1. Human DNA = 23.13 kb with λ DNA-HindIII digest (NEB)
Steps
Part A: First Elution
Heat water bath to 37°C before starting.
Thaw Buffy Coat samples by gently agitating sample in water bath.
Turn on QIAcube. Open door and remove reagent tray from QIAcube. Unscrew all caps and refill reagents where necessary. Do not exceed fill line shown on side of reagent bottle. Make sure reagents are in the proper position in reagent tray.
1. Position 1: Empty
2. Position 2: Buffer AL
3. Position 3: 96-100% Ethanol
4. Position 4: Buffer AW1
5. Position 5: Buffer AW2
6. Position 6: Buffer AE
Add a fresh set of disposable 1000 µL and 200 µL tips to QIAcube.
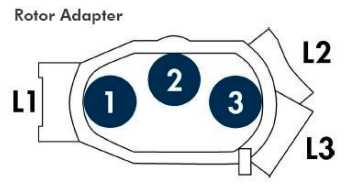
Lay out rotor adaptors on holding tray labeled DNA.
Set up rotor adaptor by placing following materials in the proper positions:
1. Position 1: QIAamp spin column
9. Lid Position: L1
2. Position 2: Empty
9. Lid Position: Empty
3. Position 3: 1.5 mL Eppendorf tube
9. Lid Position: L3
Load rotor adaptors in centrifuge of QIAcube.*
Aliquot 200µL to Sample Tubes RB taking care to pipette up as much Buffy Coat as possible.
Load Sample Tubes RB into sample tray of QIAcube.*
Aliquot 284µL into a 1.5 mL Eppendorf tube.* Place in microcentrifuge tube slot A in QIAcube.
Close door of QIAcube and begin program:
DNA > QIAamp DNA Blood Mini > blood or body fluid > DNA isolation Part A
Part B: Second Elution
Remove rotor adaptors and place on sample holding tray.
Move QIAamp spin column from 1.5 mL Eppendorf tube to Position 1 with lid in Position L1. (Part A step 6)
Remove and cap 1.5 mL tube with DNA sample and label with Freezerbondz label DNA-01.
Add a fresh 1.5 mL tube to rotor adaptor in Position 3 with lid in Position L3. (Part A step 6)
Place rotor adaptors back in the corresponding numbered position in QIAcube’s centrifuge.
Add a fresh set of 200 µL disposable tips.
Close QIAcube lid and begin program:
DNA > QIAamp DNA Blood Mini > blood or body fluid > Add Elution Part B
When program has completed, discard QIAamp spin column and remove 1.5 mL tube. Label tube with Freezerbondz label DNA-02.
Combine volume of DNA-02 to DNA-01 and pipette up and down to mix to ensure a homogenous mixture of DNA.
Split DNA back into two 200µL.
Aliquot 3µL into a 1.5 mL tube for Cary concentration assay. (Store sample in -20°C if not being immediately assayed.)
Aliquot 2µL into a PCR tube for .7% agarose gel electrophoresis to confirm for the presence and size of the DNA. (Store sample in -20°C if not being immediately assayed.)