Antibiotic treatment of the breadcrumb sponge Halichondria panicea and subsequent recolonization
Lara Schmittmann, Ute U Hentschel
host-microbe interactions
sponge model
symbioses
microbiome
metaorganism
holobiont
antibiotic treatment
Abstract
This protocol generates sponges ( Halichondria panicea ) with a disturbed microbiome under controlled experimental conditions, in order to study bacterial recolonization dynamics. Bacteria-bacteria interactions can be analysed with this set-up within the host environment aiming at a better understanding of sponge-microbe symbiosis in vivo .
It is divided into the sections 1) preparation, 2) antibiotic treatment and recovery phase, 3) recolonization with the natural microbiome and 4) sampling.
Before start
Start with section "preparation" and take ~1-2 weeks to prepare
Steps
Preparation
Autoclaving material prior to use for 15 min at 121°C
- culture bottles
- silicone tubing (in- and outflow tubes can be connected to culture bottles before autoclaving)
- 20 L carboys with tin foil wrapped around lid but don't close lid! otherwise carboys dent when they cool down
- filtration unit with inserted filter paper 0.22 µm, diameter 142 mm
- saline (1.5 % NaCl in MilliQ water)
- food stock
- CMF-ASW
Food stock
Autoclaved Nannochloropsis salina unicellular algae solution
Weigh 16 g Nannochloropsis salina powder dried algae and dissolve in 2 L MilliQ water with 30 g artificial seasalt
Shake rigourously for several minutes or use magnetic stirrer
Filter through 40 µm cell strainer to remove algae clumps
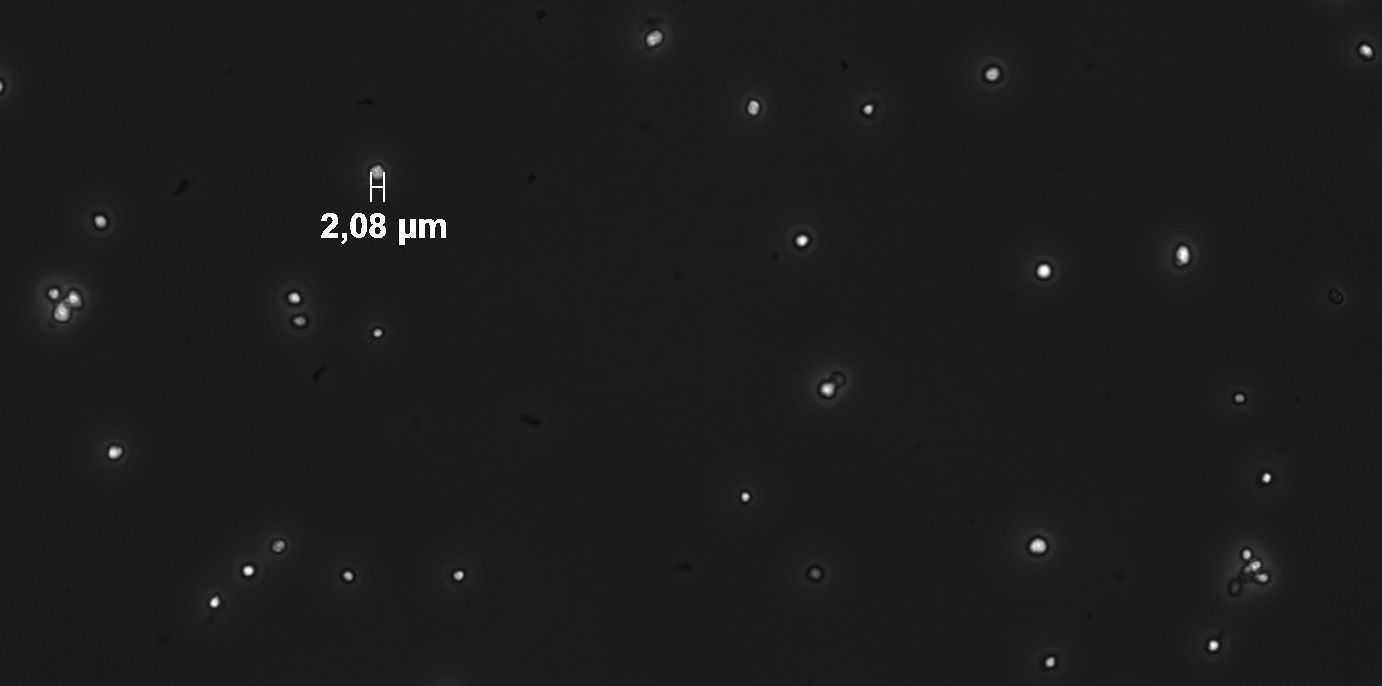
Determine concentration in stock (flow cytometer or counting under the microscope)
adjust to a stock solution of 5 x 107cells/ml (final concentration is 105cells/ml)
Autoclave in 160 mL portions
Store at RT
Antibiotic stocks
Per experiment, prepare 350 ml of each of the following antibiotic stocks:
- 50 mg/ml Rifampicin in 100 % DMSO (working conc. 50 mg/l)
- 50 mg/ml Ampicillin in MilliQ water (working conc. 50 mg/l)
- 50 mg/ml Nalidixic acid in 0.3 M NaOH (4.2 g NaOH in 350 ml MilliQ water) (working conc. 50 mg/l)
- 50 mg/ml Neomycin in MilliQ water (working conc. 50 mg/l)
- 2 mg/ml Polymixin B in MilliQ water (working conc. 2 mg/l)
Freeze in portions of 20 ml and store at -20°C
Use freshly thawed solutions, do not re-freeze
Sterile filtered artificial seawater (F-ASW)
F-ASW should be prepared every day during the antibiotic treatment and every second day during the recovery phase and recolonization
Fill a 100 l barrel with destilled water while continuously adding 1.5 kg artificial seasalt
Dissolution can be sped up by adding a water pump into the barrel to mix the water
Mix well! Check salinity at bottom and top of the barrel and prevent foramtion of a salinity gradient. Slowly add missing salt until the desired salinity is reached
Since the salinity constantly fluctuates in the Baltic Sea, we choose the ambient salinity on the starting day of each experiment (between 14 and 18 PSU)
Connect autoclaved stainless steel filter holder (142 mm) unit to pump and fill 4 autoclaved 20 l carboys with F-ASW
When all carboys are filled, stop water pump and rinse it in fresh water
Rinse filter unit and change filter paper; autoclave including silicone tube for the next filtration
Add 40 ml Nannochloropsis salina stock solution to each 20 l carboy
During antibiotic treatment, add 20 ml per antibiotic to each 20 l carboy. The final working concentrations will be:
- 50 mg/l Rifampicin
- 50 mg/l Ampicillin
- 50 mg/l Nalidixic acid
- 50 mg/l Neomycin
- 2 mg/l Polymixin B
Mix well (by rolling closed carboys)
After use, rinse carboys twice with fresh water and autoclave for the next use
MB medium
Prepare 15 g/l Difco agar and 37.5 g MarineBroth in 1 l destilled water
Prepare ~350 plates per experiment (= 10 bottles of MB medium) and store at 4°C
Cook in water bath for 10-15 min to dissolve medium
Autoclave and store at 60°C until pouring plates
Pour plates under biosafety cabinet and store upside down at 4°C
CMF-ASW (Calcium-Magnesium-free artificial seawater) after Rottmann et al. 1987
Prepare e.g. 10 l as a stock and autoclave in portions of 500 ml
For 1 l stock solution:
27 g NaCl
1 g Na2SO4
0.8 g KCl
0.18 g NaHCO3
add 1 l MilliQ water
Dilute stock 1:1 with MilliQ water and autoclave in portions of 500 ml
Can be stored at RT, but it is used ice-cold. Put bottles in freezer 1 h before they are needed
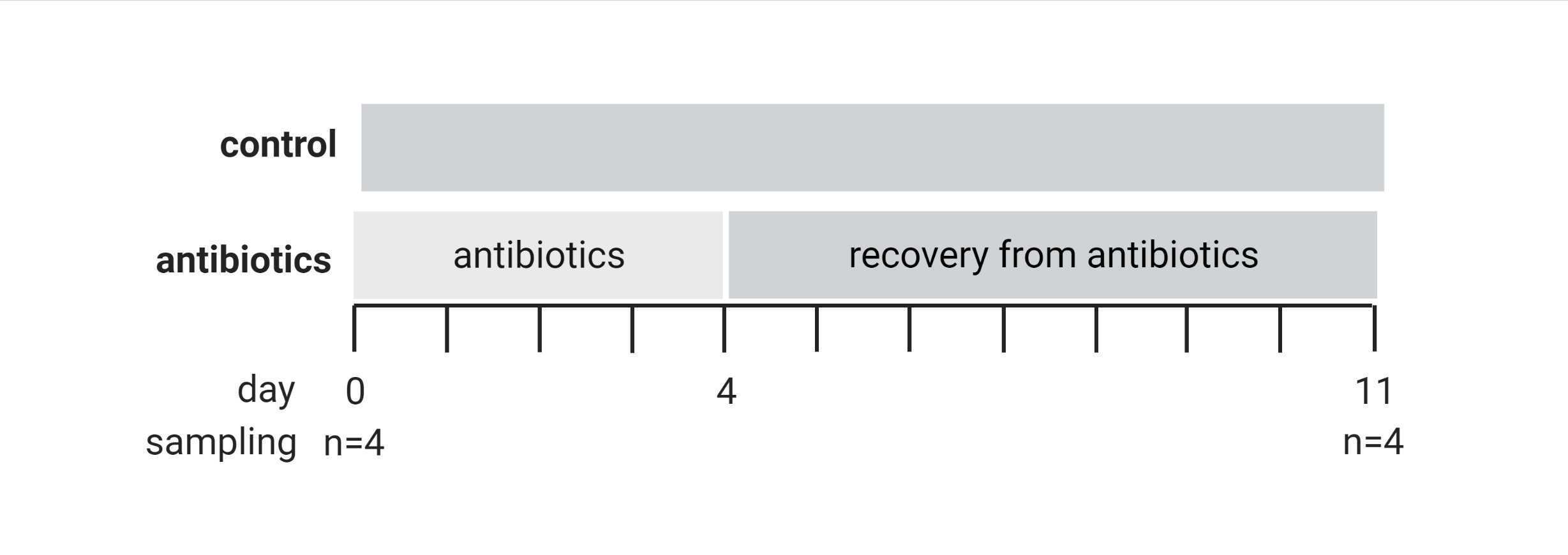
Antibiotic treatment and recovery phase
Antibiotic treatment
During the first four days of the experiment prepare ASW with antibiotics and change water source daily
Start pumps on the first day prior to placing sponges in the Erlenmeyer flasks
Set pumps to 150 doses/day of 12 ml to exchange the volume of each bottle 3.5 times per day
When Erlenmeyer flasks are filled, add one sponge in each Erlenmeyer flask (about 3x3x3 cm)
Start the experiment early in the morning
Prepare fresh antibiotic water and change carboys daily to prevent degradation and loss of function of antibiotics
Antibiotic wash-out
In the afternoon of T4 start wash-out of antibiotics by exchanging the carboys with ASW+antibiotics to carboys with ASW only
Flush the Erlenmeyer flasks several times with 250 ml fresh ASW each (by programming the pumps to dose 250 ml. Attention! The GHL2 pumps are not designed to continuously run; after each flush they need to stop for at least 15 min)
Since Rifampicin is coloured bright orange, you clearly see the reduction of antibiotics in the water by flushing
Aim at completely clear culture water until the next morning to prevent remaining antibiotics at low concentrations
It helps to manually pour out water (without opening the Erlenmeyer; simply tilt the bottle and empty it via the outflow barb) and reduce the water level to about 200 ml before flushing
Recovery phase
During the recovery phase, exchange the water source every second day with freshly filtered ASW
Set the pumps to 100 doses/day of 10 ml to exchange to volume of each bottle twice per day
Recolonization with the natural microbiome
Differential centrifugation
To prepare a bacterial inoculum for recolonization with the natural microbiome dissociate fresh, healthy sponges
The volume of sponge tissue should match the volume of sponge that you want to recolonize
Remove large pieces of algae and place sponges in a beaker with sterile, ice-cold CMF-ASW for 5 min to remove loosely attached bacteria
Resuspend the pellet in ~5 ml CMF-ASW and pool the bacteria fraction from several tubes
This is the recolonization inoculum. Keep on ice!
Proceed as quickly as possible to prevent dying of bacteria. Always keep on ice!
First recolonize, but keep a few milliliters to sample (step 12)
Replace CMF-ASW
Cut sponge tissue with a razor and forceps in a petri dish with sterile, ice-cold CMF-ASW
Only process a small piece at a time and try to keep tissue submerged
Remove algae or other non-sponge material
Transfer the pieces into 50 ml polypropylene tubes filled with ~30 ml sterile, ice-cold CMF-ASW and keep them on ice during the process
Add no more than 10 ml sponge tissue
Top up tubes until 50 ml with sterile, ice-cold CMF-ASW
Incubate tubes on ice on an orbital shaker at 200 rpm for 20 min
Filter through 40 µm cell strainer in fresh 50 ml polypropylene tubes, squeeze remaining tissue with forceps
Top up until 50 ml with sterile, ice-cold CMF-ASW
Centrifuge for 20 min at 700 g and 4°C to remove sponge cells
Transfer supernatant to a fresh 50 ml polypropylene tube and discard the sponge cell pellet
Centrifuge supernatant for 15 min at 4000 g and 4°C
Recolonization
Recolonize sponges by injection of 2 ml recolonization mix with sterile syringes and needles
Inject gradually and change location 5 times per sponge to distribute the inoculum throughout the whole animal
For the control treatment, use a sham control with 2 ml sterile CMF-ASW per sponge
Viability and composition of inoculum
Take different samples to ensure viability of inoculum and analyse bacterial composition later with qPCR and amplicon sequencing
Plate 100 µl pure inoculum, 1:10, 1:100 and 1:1000 dilution on MB agar plates at incubate at 25°C for 1 day
Check lower dilutions after 7 days
Freeze several aliquots of 1 ml pure inoculation mix in cryovials and flash freeze in liquid nitrogen
Store at -80°C
Filter 2 x 2 ml inoculum mix onto 0.22 µm filter with a reusable syringe filter holder (diameter 25 mm), store filter in cryovials and flash freeze in liquid nitrogen
Store at -80°C
Fix 2 x 1.8 ml inoculum mix with 0.2 ml Glutaraldehyde+Paraformaldehyde (final concentration 0.01 % and 1 %)
Fix for 10 min at RT
Freeze at -80°C
Repeat recolonization three times within 48 h to ensure transfer of bacteria
Sampling
Plating of culture water
To monitor bacterial growth in the Erlenmeyer flasks, culture water is plated every 2-3 days. The sterile filtered ASW and the food stock solutions should be plated before the start and at least once during the experiment to ensure sterility
Sample ~1 ml culture water from 4 replicate tanks per treatment via the silicone septum with a sterile needle and syringe. Rinse syringe once with culture water before taking the sample and take care to do it very slowly, so the sponge does not get kicked around by the water current
Store samples in fridge no longer than 3 h
Under the clean bench: prepare dilution series by pipetting 100 µL culture water and 900 µL saline (1:10) and repeat twice until a dilution of 1:1000 that is used for plating
Mix well!
Plate 100 µL of the 1:1000 dilution in triplicates on MB agar plates; use glass spatula (sterilize by dipping in ethanol and burning, cool down)
Wrap plates with parafilm and incubate upside down at 25°C for 7 days
Count colony forming units (CFUs)
Sponge tissue samples
At T0, T11 and after the recolonization take sponge tissue samples and preserve for different analyses
Take a picture of each sponge before sampling
Clean dissection place with ethanol and RNAseaway
Work quickly
Cut sponge in 6 pieces with sterile forceps and scalpel on a sterile petri dish.
Clean utensils after each sponge with ethanol and RNAseaway and discard petri dish
DNA/RNA extraction
Preserve two pieces in 2 x cryovials with 1.6 ml RNAlater
Incubate at 4°C overnight and transfer to -80°C the following day
For DNA/RNA extractions and qPCR see protocol:
Backup
Flash freeze one piece (or all leftover sponge tissue) in cryovial in liquid nitrogen and store at -80°C

