Acquiring and integrating data from multiple resources for meta-analysis
Wu Huang, Alex Twyford, Peter Hollingsworth
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
Acquiring sequence data generated for various purposes and repurposing it to evaluate the nature of genomic differences between plant species requires thorough dataset assessments, efficient data processing, and careful management. In this chapter, I summarise the methodology for searching appropriate datasets, data collection and filtering, and data management. I provide examples of the data formats to guide future downloading and acquiring of the datasets, and the links to curated publicly available bioinformatic tools and self-written scripts to allow reproduction and reuse of this data acquiring and processing pipeline.
There is now a significant number of studies that have sequenced multiple loci from the nuclear genomes of plants and these are available in public databases or in private data repositories (if the study hasn’t yet been published). Collectively these datasets have great potential for understanding the nature of genomic differences between plant species. To harness this existing information I have developed a set of workflows, with the logic behind my approach being to
● Develop key criteria for selecting suitable datasets focusing only on datasets which have sampled multiple individuals from multiple congeneric species
● Search the literature and public data repositories for datasets that match these criteria
● Obtain and organise the selected datasets
● Use these datasets to estimate the proportion of species that resolve as monophyletic units as one measure of species discrimination success
● Establish the frequency distribution of taxonomically informative nucleotide substitutions among taxa to better understand the genomic nature of differences between plant species
● Subsample the data to better understand the effectiveness of smaller numbers of loci in recovering maximal species
discrimination, as well as evaluating the attributes of the gene regions that are most effective at telling species apart.
This protocol will focus on the first three bullet points. The protocol for the rest of the bullet points please refer to another protocol "NucBarcoder - a bioinformatic pipeline to characterise the genetic basis of plant species differences".
Steps
Developing data request form
Sourcing the literature and compiling publications which sequenced multiple nuclear loci for multiple individuals from multiple congeneric species.
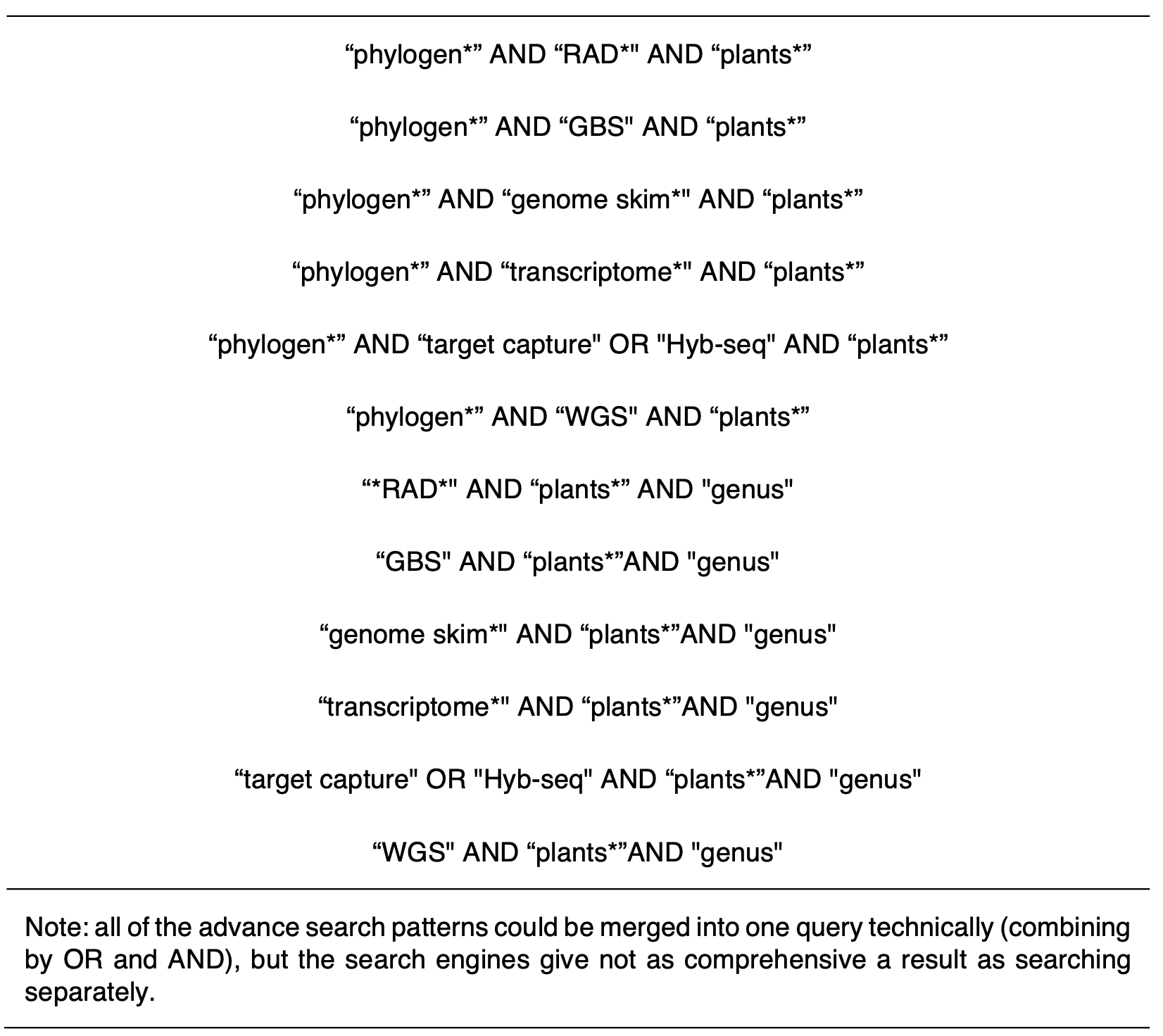
To compile data suitable for meta-analysis, We first searched journal publications from 2013 onwards for studies that sequenced multiple loci from the nuclear genome and which sampled multiple individuals of multiple congeneric species. The cut-off of 2013 was selected as this reflects the initiation of widespread use of next-generation sequence platforms for the recovery of nuclear sequence data from plants. We used ambiguous matching patterns to search in the Web of Science and University of Edinburgh literature search engines. The matching patterns are listed in Table 1.1
With the full-text publications downloaded, the next step was selecting publications manually.
I only kept studies if they satisfied the following criteria.
1) Three or more un-linked nuclear loci were sequenced.
2) More than two species had multiple individuals successfully sequenced and retained.
3) A phylogeny is available either in visual format or in a machine readable text format (newick, phylip, or nexus formats).
4) The species identities on the phylogeny could be interpreted and related to the sampled species.
Reading the methods and concluding the two popular NGS analysis pipelines are HybPiper and iPyRAD. Analyse the files used in each step and evaluate the pros and cons of acquiring data from that step.
| A | B | C | D | E | F |
|---|---|---|---|---|---|
| Data_stages | Features_of_the_stage | Pros | Cons | File_format | Note |
| Clean_reads | The original data | 1. Intact information, can recover potential heterozygous sites, sequencing error, and coverage | 1. Big files (10 Mb * N) 2. Parsing reads file needs comprehensive computational analysis 3. Need extra step to analyse if there are samples to exclude | fastq | file size denote a relative number N - number of samples |
| Cluster reads by alignment | Clustered and aligned reads data to the reference sequence (bwa). Will filter reads according to standards. | discarded putative contaminations remained potential heterozygous sites, sequencing error, and coverage info | 1. Big files (4 Mb * N) 2. Computationally expensive (can easily generate vcf file) 3. Need extra step to analyse if there are samples to exclude | bam | I think this file as well as the reference_gene.fasta would be the best choice. |
| Sequence Assembly | Assembled each cluster in each samples | Small files (0.003 Mb * N) | 1. Lost heterozygous information. No sequence error info, no coverage info. Still need alignments 2. Need extra step to analyse if there are samples to exclude | fasta | |
| Multi-align assembled sequence | Align all the assembles sequence together | 1. Small files (0.003 *N Mb) 2. Easy to analyse | Lost heterozygous information. No sequence error info, no coverage info. | fasta | I am currently use this for Inga and Geonoma analyses |
| Variant calling file | Variation called | 1. Small files (0.001 *N Mb) 2. Easy to analyse | Lost heterozygous information. No sequence error info, no coverage info. No non-variant sites info | fasta |
Table 2.1. Pros and cons of acquiring data from the main steps of HybPiper.
Collecting information about the file formats frequently used in the data repositories that are popular in this particular field. (Normally read >20 related publications and the picture will be clear.) Choose the most frequently used ones as input formats for the following analytical pipelines. Three important files are:
Metadata. This includes information that corresponds to the sample IDs in the consensus sequence files which links sequences of individuals to their scientific names (species identities). This information is also useful for tracking changes of names wherever this has been applied, as well as monitoring and understanding the inclusion and exclusion of individuals and loci.
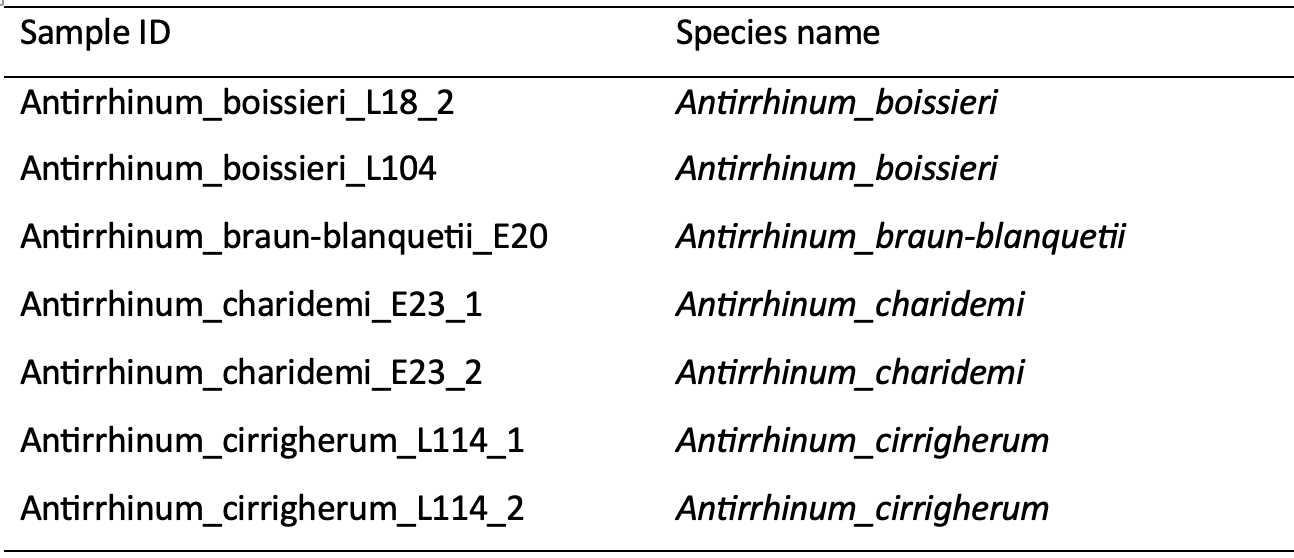
Sequence alignment file. This alignment covers the sequence variation in all individuals per dataset, and depending on how the original raw reads were processed, takes the form of either multiple-aligned sequences in .fasta, .phylip, or .nex formats, or SNP matrix in .vcf or .fasta format.

Phylogenetic trees. Phylogenetic trees were recovered from each study to enable easy estimation of the proportion of species that resolve as monophyletic. The preferred format is for this to be a machine-readable text format such as the Newick format. However, where only a graphical representation of the tree was available, this was also retained and used, to maximise the number of studies analysed.

Acquiring data
Through the literature review, identify published data that could be reused.
Email the corresponding author of published data asking for their consent of using their data (though it's always free to use published data, it helps to build rapport and potentially will gain additional insights into the data used). Attached is an example of the email I used for this request. Cold email requesting data reuse.docx
Extract metadata from the publication manually.
Download data from the data repositories, such as Zenodo and Dryad. Email the correspondent author for required data or metadata if the files are not self-explanatory. The chance of having a helpful reply is low though.
In addition to mining the published literature, I contacted potential collaborators to request access to unpublished datasets. This involved designing a data request form and developing a list of metadata entries. This is for standardising metadata and agree with a data-sharing scheme. Meta-data_info.xlsx
Identify potential working groups that have unpublished data. Cover letter.docx
Ask people to fill in the metadata entries.
Send them the guidance for sharing files, including what files to share, where to share, and how the data will be used and authored. Requesting data form.docx
Data storage and backup
The shared data were uploaded to Google Drive (https://www.google.co.uk/intl/en-GB/drive/), and then downloaded and managed on the UK crop diversity bioinformatics HPC platform (https://www.cropdiversity.ac.uk).
Two copies of the backup are continually updated. A full version of data is backed up on the local hard drive and an abridged version ison the laptop.

