Accurate profiling of diazotrophic communities using unique molecular identifiers with Nanopore sequencing
Nobuhiko Shigyo
Abstract
This protocol applies the method by Karst et al. (2021), who used UMI-tagged primers to perform highly accurate long-read amplicon analysis on the full-length ribosomal RNA operons, to the nitrogen fixation ( nif ) gene cluster. A recent study has shown that the functional gene nifH , which is frequently used in community analysis of diazotrophs, detects many false positives (Mise et al. 2021). Therefore, in this protocol, I used newly developed primers targeting the nifD-K gene (approx. 1.8 kbp) to identify the diazotrophic communities. The present protocol is presented using plant litter and mineral soil as examples, but it could potentially be applied in a variety of environmental samples.
Steps
DNA extraction
Extract total genomic DNA (gDNA) from 500mgof fresh soil and 100mg of air-dried litter using the ISOIL for Beads Beating kit (Nippon Gene Corporation, Tokyo, Japan), a CTAB-based DNA extraction kit.
Measure the extracted gDNA concentration using a Qubit fluorometer with the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Waltham, MA, USA).
Tagging nifD-K genes with UMIs (1st PCR)
Prepare PCR-master mix in a 0.2 mL PCR tube with the following:
| A | B |
|---|---|
| Nuclease-free water | X µL |
| Forward primer (nifDK229f_UMI, 10µM) | 2.5 µL |
| Reverse primer (nifK476r_UMI, 10 µM) | 2.5 µL |
| PrimeSTAR Max Premix | 25 µL |
| Template DNA | Y µL (50 ng) |
Run the following PCR program:
[98℃ 10 sec → 55℃ 15 sec → 72℃ 30 sec] × 2 → 8℃ ∞
Clean-up of 1st PCR products
Clean up the 1st PCR products using KAPA HyperPure Beads (Kapa Biosystems, Inc., Wilmington, MA, USA)
Equilibrate the beads solution at room temperature and allow the beads to resuspend completely.
Dry the beads at room temperature for 3 min.
Remove the tube from the magnetic rack.
Elute the purified DNA by adding 21µL of nuclease-free water and mix by pipetting.
Incubate the tube at room temperature for 5 min.
Place the tube on the magnetic rack to capture the beads. Incubate until the liquid is clear (1 min).
Transfer the supernatant to a new PCR tube.
Add 25µL beads solution to the 50µL PCR products (0.5× bead solution/sample ratio) and mix by pipetting up and down multiple (10 to 20) times.
Incubate the tube at room temperature for 5 min.
Place the tube on a magnetic rack (NGS MagnaStand (YS-Model) 8Ch × 0.2 mL PCR tube, Nippon Genetics Co., Ltd., Tokyo, Japan) to capture the beads. Incubate until the liquid is clear (~3 min).
Discard the supernatant.
Wash beads by adding 200µL fresh 80% ethanol.
Incubate the tube on the magnetic rack at room temperature for 30 sec.
Discard the supernatant.
Repeat the previous washing steps (4.6-4.8).
Amplification of UMI tagged sequences (2nd PCR)
Prepare PCR-master mix in a 0.2 mL PCR tube with the following:
| A | B |
|---|---|
| Nuclease-free water | 20 µL |
| Forward primer (lu_pcr_i1_fw_v7) | 5 µL (0.5 µM) |
| Reverse primer (lu_pcr_i1_rv_v7) | 5 µL (0.5 µM) |
| PrimeSTAR Max Premix | 50 µL |
| Template DNA (1st PCR products) | 20 µL |
Run the following PCR program:
[98℃ 10 sec → 68℃ 5 sec → 72℃ 30 sec]×30 → 68℃ 5min → 8℃ ∞
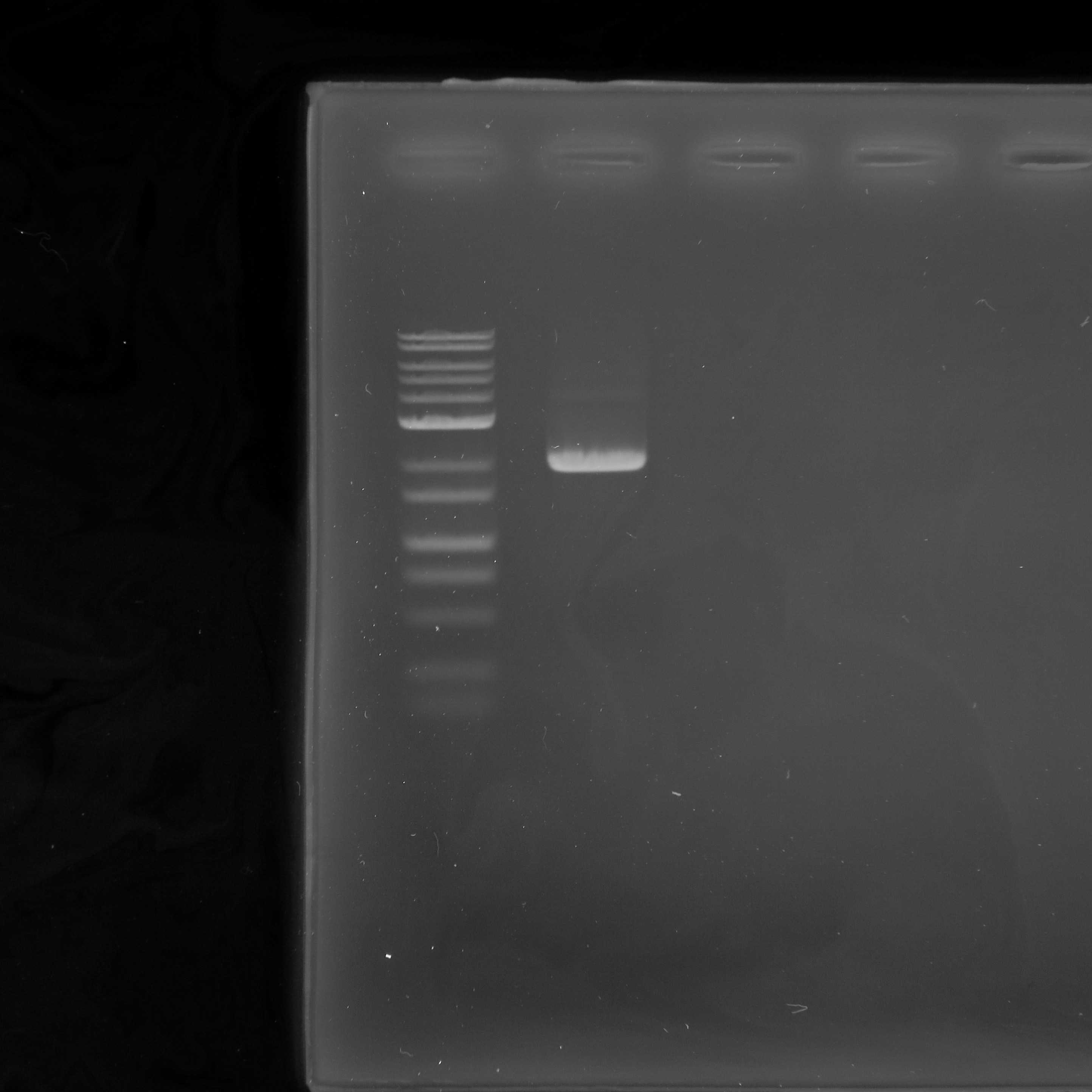
Clean-up of 2nd PCR products
Clean up the 2nd PCR products using KAPA HyperPure Beads (0.5× bead solution/sample ratio) following the same procedure as in Section 5.
Quality control
Measure the DNA concentration of the purified 2nd PCR products using a Qubit fluorometer with the Qubit dsDNA HS assay kit.
Nanopore sequencing
Conduct "DNA repair and end prep", "Adapter ligation and cleanup", and "Priming and loading of SpotOn flow cell" according to the Ligation Sequence Amplicon V14 protocol (ligation-sequencing-amplicons-sqk-lsk114-ACDE_9163_v114_revE_29Jun2022-minion.pdf).