A versatile nuclei extraction protocol for single nucleus sequencing in non model species – optimization in various Atlantic salmon tissues.
Rose Ruiz Daniels, Richard S Taylor, Ross Dobie, Sarah Salisbury, Emily Clark, Dan Macqueen, Diego Robledo
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
Single cell RNA sequencing has rapidly become a standard tool for profiling transcriptomic diversity across thousands of cells (Linnarsson and Teichmann, 2016), and is now being applied to a large diversity of species and tissues. The main limitation of this technology is that it requires the isolation of live cells from fresh tissue, severely restricting its applicability. As a result, single nuclei RNA sequencing (snRNA-seq), which consists of sequencing the RNA of only the nuclei of cells rather than of the whole cell, has been commonly adopted since it allows samples to be stored for several months prior to processing while yielding comparable results to whole cell sequencing (Kulkarni, et al., 2019; Slyper et al. 2021). A critical challenge for snRNA-seq is the successful extraction of high quality nuclei. This has spurred the recent publication of a number of dissociation protocols for nuclei extraction (Drokhlyansky et al. 2020; Eraslan et al. 2021; Melms et al 2021), however, these have largely been optimized for model species such as humans, and more and more single nuclei is being adopted in non-model species.
Here we present a robust protocol that enables the extraction of nuclei from frozen tissue adapted from those shown to work in different tissue types, such as human skin (Drokhlyansky et al. 2020; Eraslan et al. 2021; Melms et al 2021). Our protocol has been used to successfully extract nuclei from an array of different Atlantic salmon (Salmo salar) tissues including liver, skin, fin, spleen, head kidney and gill as well as in other species such as sole (Solea solea) nose and gonad, rabbit (Oryctolagus cuniculus) nasal tissue and nurse shark (Ginglymostoma cirratum) spleen. We present the protocol as applied to fin and skin as these are particularly challenging tissues to work with given their toughness and the presence of hard tissue (e.g., scales and bones), connective tissue and fat deposits. We include notes throughout the protocol so that the reader can optimise it for a variety of tissue types. While the protocol has been optimised to work with the Chromium 10x platform, the most commonly used high throughput microfluidic device, but can be used successfully for the extraction of nuclei for other platforms and applications. The aim of this protocol is to capture 7,000 nuclei per single-nuclei RNA sequencing library using the Chromium Single Cell 3’ Reagent Kits v2 or v3 (10X Genomics). Given its utility for isolating nuclei from difficult to dissociate tissue types, we anticipate that this protocol will be broadly applicable for snRNA-seq of non-model organisms and unconventional tissue types.
Before start
Sampling and storage for nuclear isolation.
Animals must be appropriately euthanized and immediately processed. Approximately ~60mg of tissue is placed in one clearly labelled cryotube and immediately flash frozen in liquid nitrogen. This step is critical . The tissue must be preserved as fast as possible for optimal results. In the absence of liquid nitrogen, samples can be frozen in dry ice. Samples can be stored at -80°C for up to a year prior to use. Older samples might still yield viable nuclei but this would need to be tested.
All reagents should be chilled on ice prior to use.
Samples should be kept frozen on dry ice until immediately before nuclei isolation, and all sample-handling steps should be performed on ice.
The centrifuge should be pre chilled at 4°C.
All reagents are given for 2 nuclear isolations.
Amounts of buffer especially those that contain RNase should be adjusted appropriately for each experiment prepared prior and RNase added immediately before use.
Before using this prep for library preparation do a trial run.
Recommended to do a trial especially on a new tissue type to adjust different parameters without adding RNase. Once parameters are adjusted such as mincing times, filter size and dilution in to final buffer in order to get good quality nuclei.
Attachments
Steps
Nucleus isolation workflow for ST-based buffers
On ice, place a piece of frozen tissue into one well of a 6-well tissue culture plate with 1mL TST.
On ice, mince tissue initially using Tungsten Carbide scissors for 0h 0m 30s and then with Noyes Spring Scissors 0h 10m 0s.
0h 5m 0s into the mincing gently pipette up and down with a p1000 pipette using a low retention filtered tip. The time in the dissociation buffer is critical. See image for how to assess the timing is correct by looking at your nuclei.
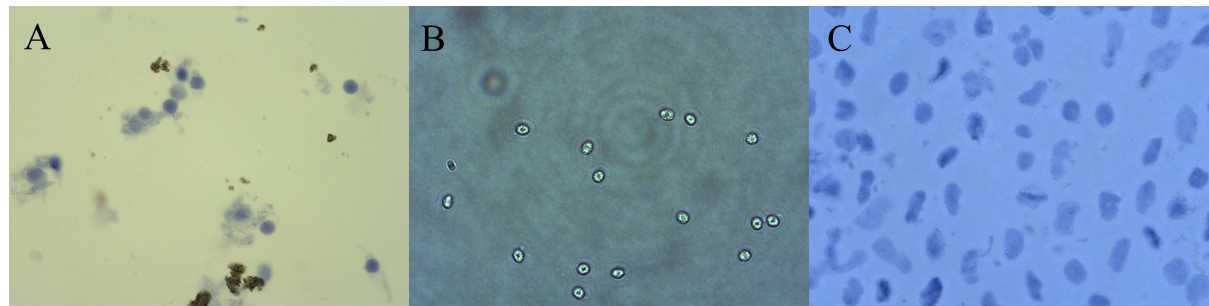
Pass lysate through a 40 µm cell strainer .
Add a further 1mL of TST to the cell strainer immediately.
Add 3mL of freshly prepared ST buffer to the lysate.
Add the 5mL of lysate to a marked 15 ml falcon tube (Corning) on ice.
Centrifuge at 500x g,4°C in a swinging bucket centrifuge.
Resuspend the pellet gently using a p1000 pipette in PBS-BSA.
Filter the nucleus solution a second time.
Count the nuclei using a C-chip disposable haemocytometer.
The nuclei are also counted using a Bio-Rad TC20 to confirm results from the disposable haemocytometer and to count the proportion of viable cells.
Load the nucleus suspension into a Chromium Chip and into the Chromium Controller, aiming to recover 7,000 nuclei as per 10x recommendations with a concentration of between 700 to 1200 nuclei per µl.

