A universal protocol for high-quality DNA and RNA isolation from diverse plant species
Farhad Masoomi-Aladizgeh, Leila Jabbari, Reza Khayam Nekouei, Ali Aalami, Brian J Atwell, Paul A Haynes
Abstract
Next-generation sequencing demands high-quality nucleic acid, yet isolating DNA and RNA from plant tissues is often challenging. Despite advancements in the development of a variety of kits and reagents, these products are only limited to isolating nucleic acid from model plant species. In this study, a universal lysis buffer is introduced to isolate nucleic acid from a wide range of plant species to facilitate molecular studies, such as quantitative PCR (qPCR), transcriptomics, whole-genome sequencing, etc. The lysis buffer consists of hexadecyltrimethylammonium bromide (CTAB), sodium chloride (NaCl), tris base, ethylenediaminetetraacetic acid (EDTA) and β-mercaptoethanol (βME). The appropriate concentration of these components enables high-quality DNA and RNA isolation from plant tissues simultaneously. Slight alterations in the methodology lead to the isolation of pure DNA or RNA from the same tissue, enabling high-throughput sequencing. This protocol is fast and enables the isolation of nucleic acid from diverse plant species in less than an hour.
Steps
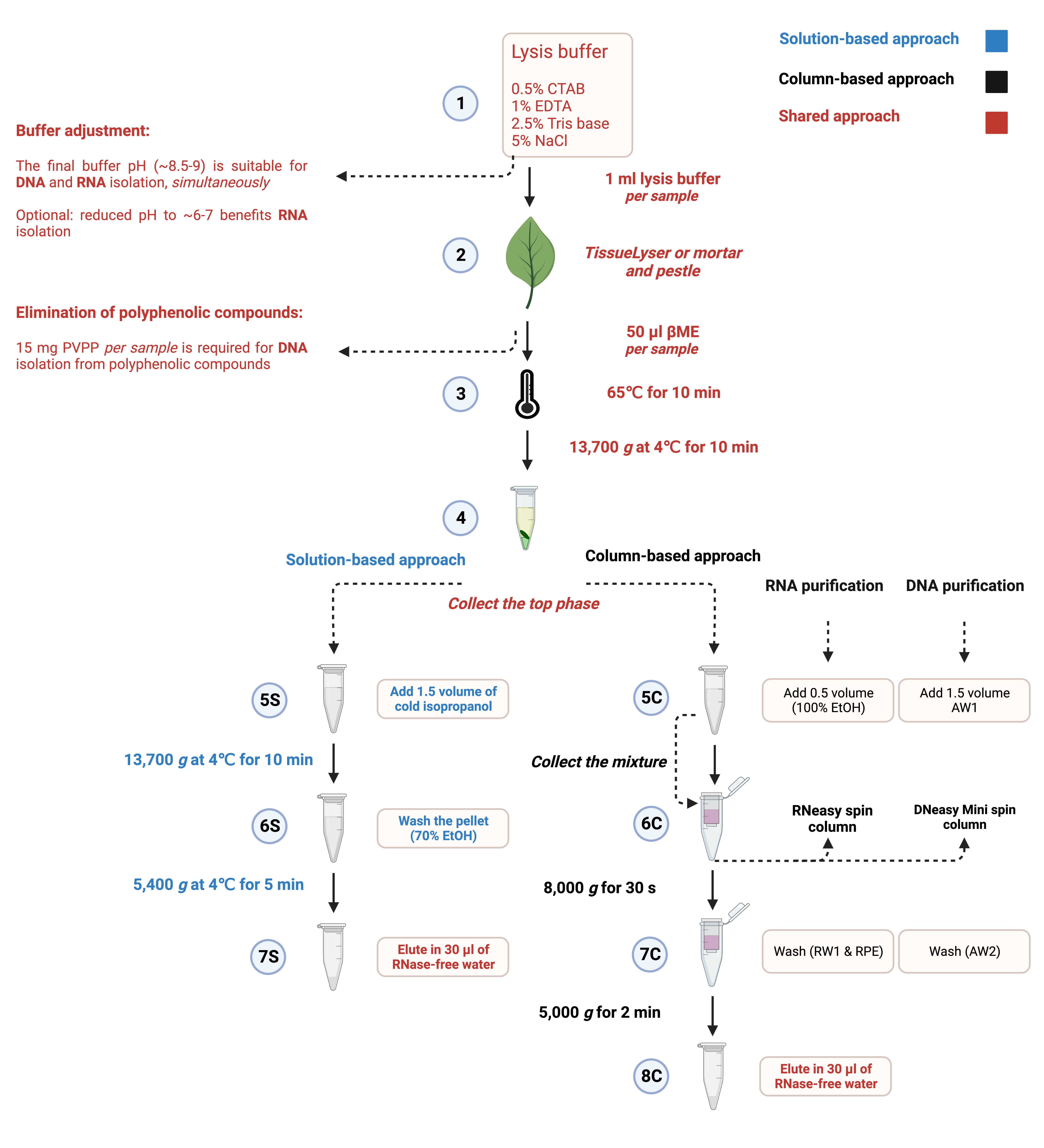
Lysis Buffer for DNA and RNA Isolation
The lysis buffer contains 0.5% CTAB, 1% EDTA, 2.5% Tris base and 5% NaCl. These are the four main components of the lysis buffer for DNA and RNA isolation from plant tissues.
Example: Add CTAB (125 mg), EDTA (250 mg), Tris base (625 mg), and NaCl (1250 mg) to 25 ml nuclease-free water, and then dissolve the components in the given order by shaking at room temperature.
CRITICAL STEP 1: PH adjustment of the lysis buffer
Option 1: The lysis buffer's pH is already 8.5-9, simultaneously isolating DNA and RNA from the mixture.
Option 2: Reducing the pH level of the mixture to 6-7 enables the isolation of only RNA from the mixture. The acidic lysis buffer precipitates DNA into the organic phase.
CRITICAL STEP 2: Supplements to the lysis buffer
β-mercaptoethanol (βME): For DNA and RNA isolation, add 50 µl (5%) βME to 1 ml of the lysis buffer before use to decrease the probable oxidation only for tissues with high polysaccharides and secondary metabolites. βME is highly recommended for RNA isolation since it eliminates RNAses released during cell lysis.
Polyvinylpolypyrrolidone (PVPP): For DNA isolation, add 15 mg (1.5%) PVPP per 1 ml of the lysis buffer only for tissues with high polyphenolic compounds to increase the concentration of DNA.
Homogenization of Tissues
Grind plant tissues (leaf, shoot, root, etc., approximately 0.1 g) following one of the below options:
Option 1: Grind the tissues with 1 ml of the lysis buffer in a sterilized mortar and pestle without liquid nitrogen.
Option 2: Grind the tissues with a TissueLyser using up to six Zirconox beads (2.8–3.3 mm) and liquid nitrogen. Then, mix the ground tissue with 1 ml of the lysis buffer, followed by vigorous vortexing to create a homogeneous mixture.
CRITICAL STEP: βME can be added to the homogenised plant tissues at this stage. Alternatively, it can be added directly to the lysis buffer as described in the first section. The former is recommended as it can be easily managed inside a chemical fume hood.
Triple-Phase Separation
Incubate samples at 65℃ for 10 min to lyse cells completely. Invert the mixture several times during the incubation time or use a thermo shaker incubator with constant shaking at 300 rpm.
Add 600 μl Chloroform to each sample and homogenize the mixture by vortexing vigorously, followed by centrifugation at 13,700 g at 4℃ for 10 min.
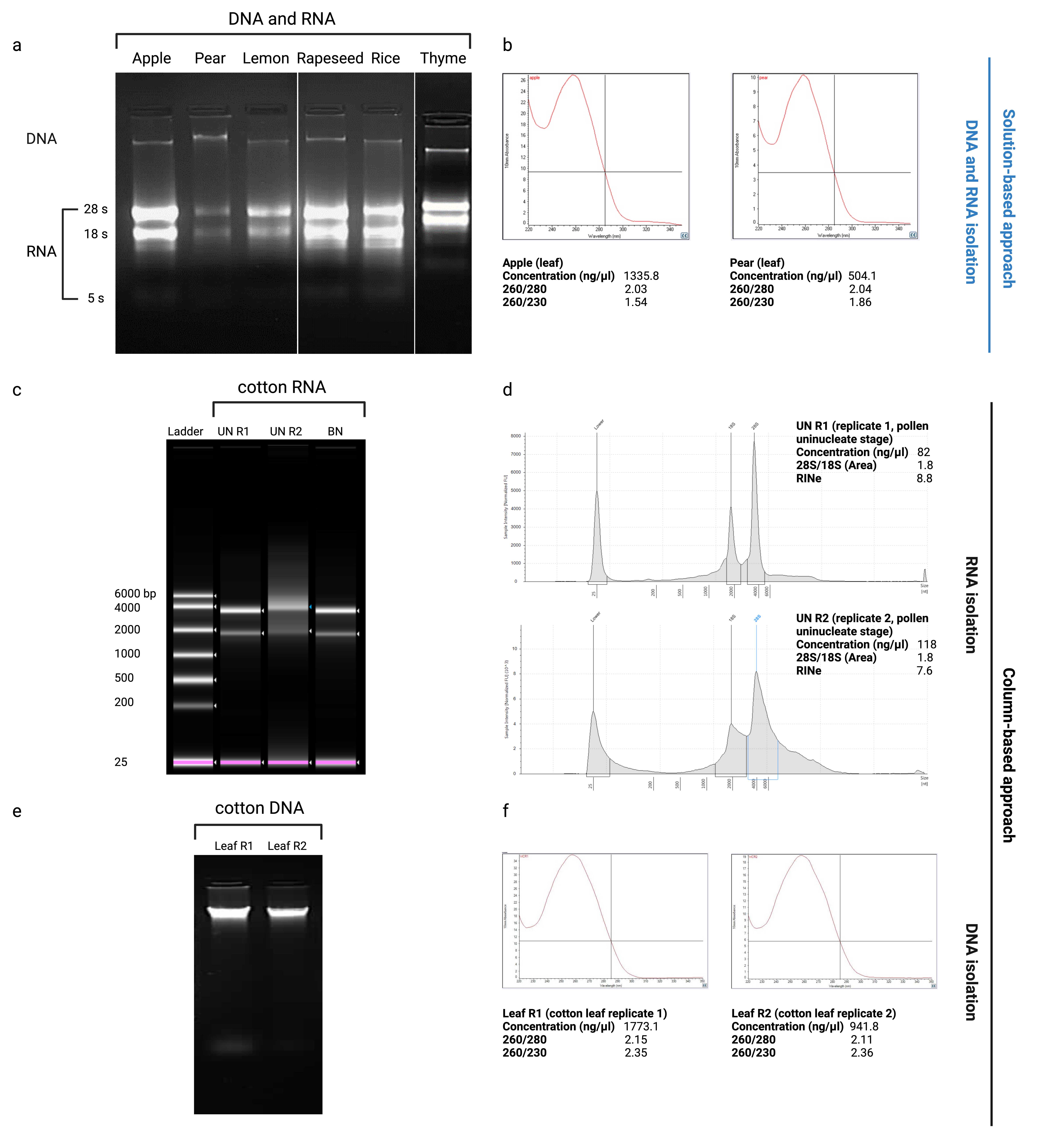
The centrifugation of the mixture results in three phases, as shown in Figure 1:
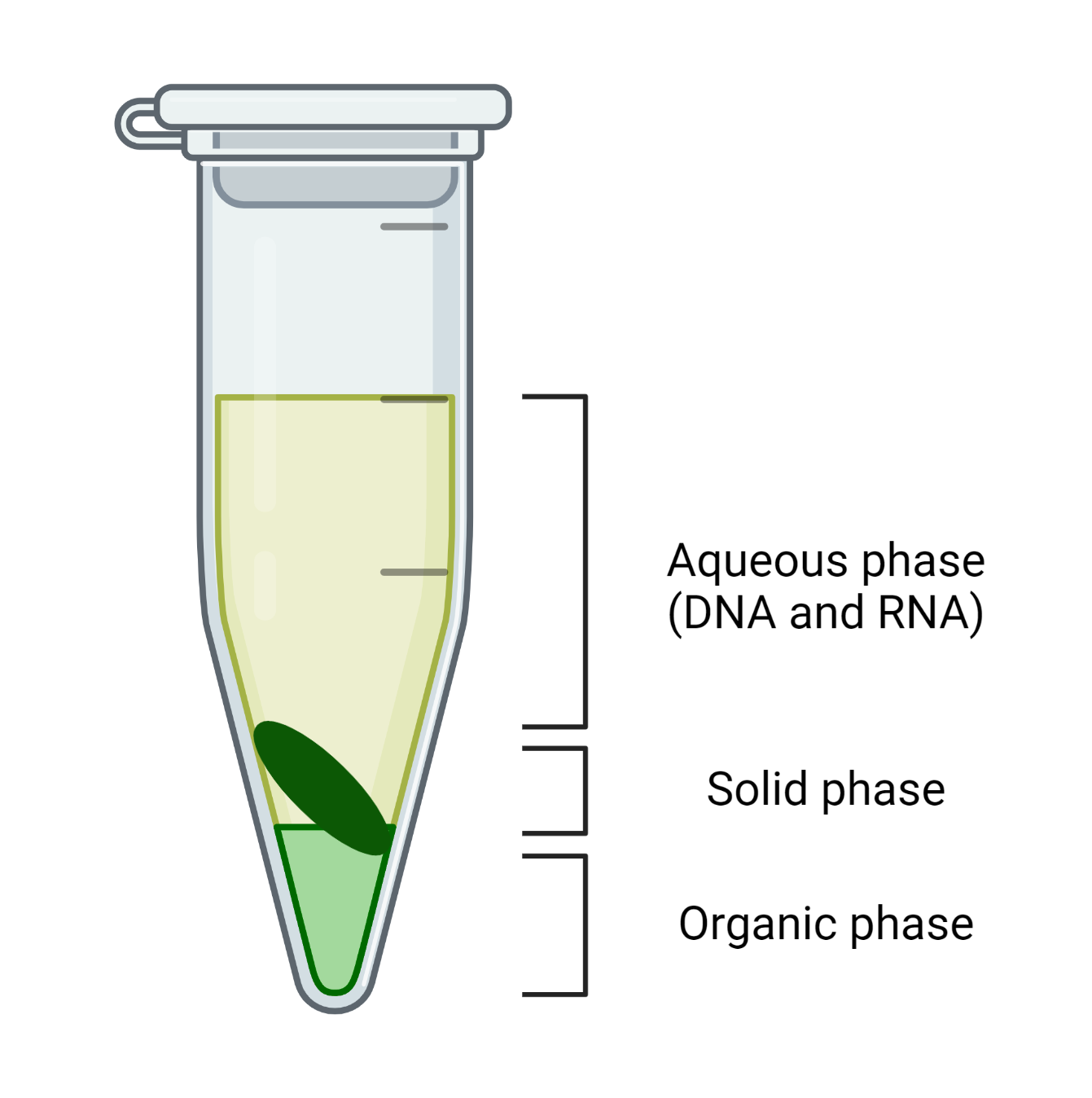
Transfer the upper aqueous phase (600 µl) to a new 2 ml microcentrifuge tube gently, without disrupting the solid and organic phases. This phase can undergo solution-based or column-based approaches for nucleic acid purification and precipitation.
CRITICAL STEP 1: The upper aqueous phase with the isolation buffer at pH 8.5-9 consists of DNA and RNA, while the isolation buffer at pH 6-7 consists of RNA since DNA participates at an acidic pH.
CRITICAL STEP 2: Continue the remaining steps at cool conditions, handling samples on ice and centrifugation at 4℃, only if RNA isolation is considered.
Purification and Precipitation of Nucleic Acid (solution-based approach)
The following procedure describes a solution-based approach for the purification and precipitation of nucleic acid, which is recommended for routine PCR, qPCR and Sanger sequencing applications.
Collect the upper aqueous phase (600 µl), add 1.5 volume (900 µl) of cold isopropanol to the mixture and invert the tube gently to mix the solution. A stringy white mass might be visible at this stage representing nucleic acids.
Centrifuge the samples at 13,700 g at 4℃ for 10 min. The white pellet will be visible on the bottom of the tubes.
Discard the supernatant and wash the pellet gently with 70% EtOH.
Centrifuge the samples at 5400 g at 4˚C for 5 min, remove the supernatant and then air dry the pellet.
Dissolve the pellet in 30 µl of RNase-free or autoclaved water by incubating at room temperature. Dissolving the pellet should take only a few minutes if the extracted DNA or RNA is pure.
DNAase I and RNase I can be used to eliminate DNA and RNA, respectively, according to the manufacturer's instructions.
Purification of and Precipitation Nucleic Acid (column-based approach)
The following procedure describes a column-based approach for the purification and precipitation of nucleic acid, which is recommended for Next Generation Sequencing applications, such as DNA-seq or RNA-seq.
DNA isolation (coupled with the Qiagen DNeasy Plant Mini Kit):
Collect the upper aqueous phase (600 µl), add 1.5 volume of buffer AW1 to the lysate, and mix it immediately by pipetting. Do not centrifuge the lysate at this stage. For example, to 600 μl supernatant, add 900 μl buffer AW1.
Transfer the mixture (~ 600 μl each time) into a DNeasy Mini spin column (white) , including any precipitate. Centrifuge the samples at 8,000 g for 30 s, and discard the flow-through. Repeat the step with the remaining lysate.
Place the DNeasy Mini spin column into a new 2 ml microcentrifuge tube, add 500 μl buffer AW2 and centrifuge at 8,000 g for 30 s. This step may be repeated more than once, especially if the spin column membrane is not clean.
Transfer the DNeasy Mini spin column to a 1.5 ml microcentrifuge tube, and pipet 30 μl RNAse-free water directly onto the DNeasy membrane. After about 5 minutes, centrifuge at 5,000 g for 2 min to elute DNA.
To eliminate any remaining RNA, treat the samples with 2 μl RNase I at room temperature for 10 min.
RNA isolation (coupled with the Qiagen RNeasy Plant Mini Kit):
Collect the upper aqueous phase (600 µl), add 0.5 volume of ethanol (96–100%) to the lysate, and mix it immediately by pipetting. Do not centrifuge the lysate at this stage. For example, to 600 μl supernatant, add 300 μl ethanol.
Transfer the mixture (~ 600 μl each time) into an RNeasy spin column (pink) , including any precipitate. Centrifuge the samples at 8,000 g for 30 s, and discard the flow-through. Repeat the step with the remaining lysate.
Place the RNeasy spin column into a new 2 ml microcentrifuge tube, add 700 μl buffer RW1 and centrifuge at 8,000 g for 30 s.
Add 500 μl buffer RPE to the RNeasy spin column and centrifuge at 8,000 g for 30 s to wash the spin column membrane (ensure that ethanol is added to the buffer). This step may be repeated more than once, especially if the spin column membrane is not clean.
Transfer the RNeasy spin column to a 1.5 ml microcentrifuge tube, and pipet 30 μl RNAse-free water directly onto the RNeasy membrane. After about 5 minutes, centrifuge at 5,000 g for 2 min to elute RNA.
To eliminate any remaining DNA in the samples, treat the samples with DNAase I according to the manufacturer's instructions.
Storage Condition
Nucleic acids can be stored either at −20˚C for short time storage or at −80˚C for longtime storage.
Workflow for Nucleic Acid Isolation
Expected Results