A method for RNA extraction, cDNA synthesis and and quantitative PCR from NIH-3T3 fibroblasts
Suzanne R Pfeffer, Sreeja V Nair
Abstract
Here we describe a protocol for RNA extraction, cDNA synthesis and quantitative PCR from NIH-3T3 fibroblasts. This method has also been used with mouse embryonic fibroblasts.
Steps
RNA extraction, cDNA synthesis and and quantitative PCR from NIH-3T3 fibroblasts
RNA extraction
Plate 0.3 X 106 cells NIH-3T3 fibroblasts or MEF cells in individual wells of a 6-well plate.
Repeat steps 8-9
Remove the supernatant using a 1 mL pipette. Decant the remaining volume onto a Kimwipe tissue. A RNA pellet should be visible at the bottom of the tube. Care should be taken to avoid losing the RNA pellet while removing the supernatant. Allow the RNA pellet to air dry.
Resuspend the pellet in 50µL RNase-free water and check the quality and concentration of RNA by Nanodrop. Purity of RNA is determined by the ratio of absorbances at 260 nm to 280 nm. A ratio >2 is considered ideal.
Store the RNA at -80°C or proceed to cDNA synthesis as described below.
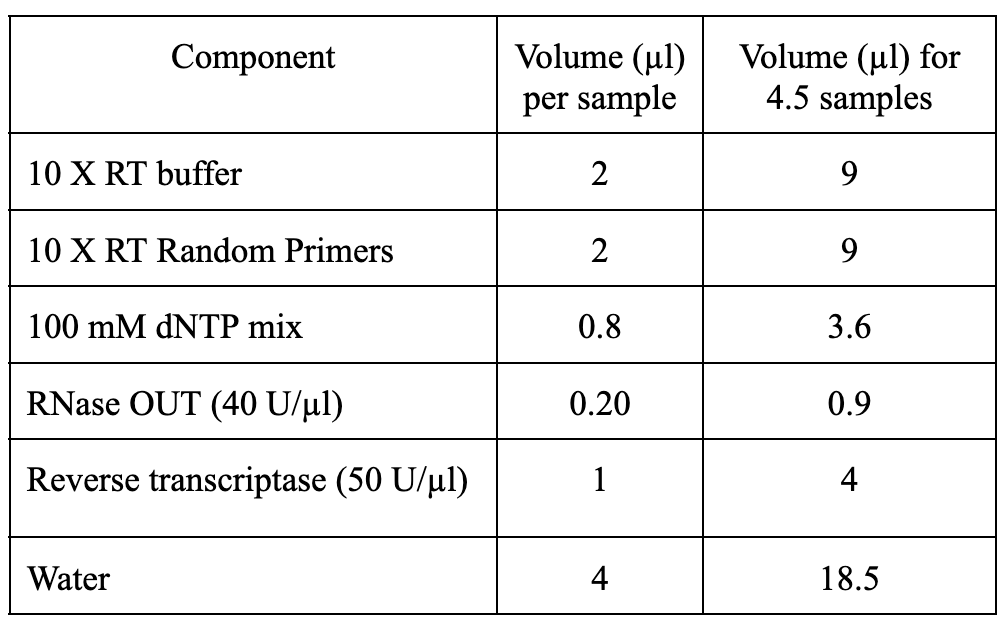
cDNA synthesis using High-Capacity cDNA Reverse Transcription Kit
- Briefly spin the tube and add
10µLof the Master Mix to labeled PCR tubes for each sample (4 tubes here) onOn ice.
- Add
500ngRNA (calculate the volume required for 500 ng from the concentration of RNA extracted) to each tube and bring the total reaction volume to20µLwith sterile waterOn ice.
- Tap the tubes once or twice to mix and set up the cDNA synthesis reaction in a PCR machine as follows: 25°C-10 min, 37°C-120 min, 85°C-5 min, 4°C-hold. Store the samples at
-20°Cor proceed to qPCR reactions on ice as described below.
- Store the samples at
-20°Cor proceed to qPCR reactions on ice as described below.
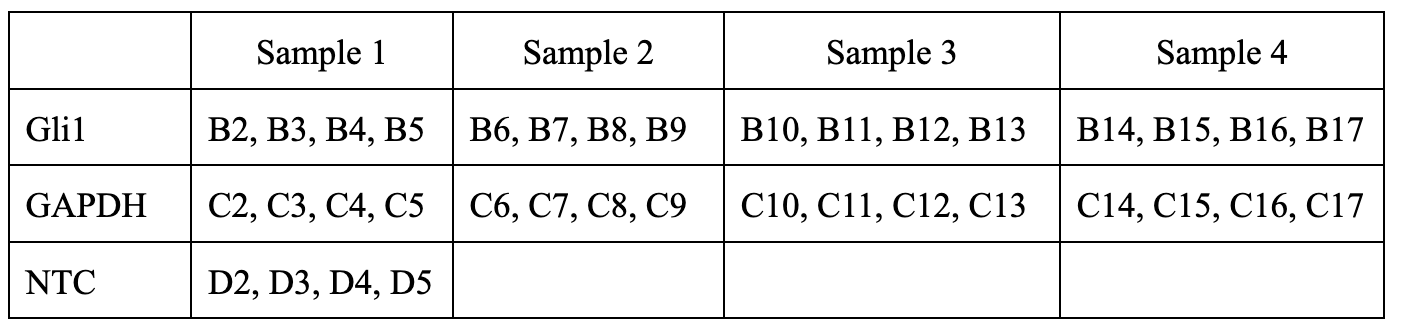
Quantitative PCR
Dilute the cDNA 20 fold in sterile water.
Add 0.5 mL Trizol reagent to each well and incubate for 0h 5m 0s at Room temperature. Scrape the cells with a 1mL pipette tip during the 5 min incubation, to help cell lysis.
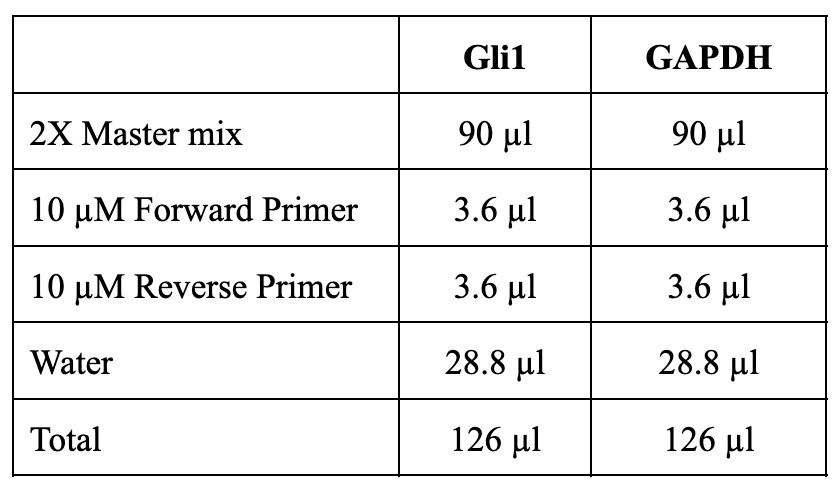
Prepare a 2X master mix for each RNA to be analyzed. Here, as an example, we are analyzing Gli1 levels in each sample with GAPDH as an internal control. Calculations are shown below:
Seal the plate, centrifuge the plate briefly to collect the solutions to the bottom of the well. Set the PCR reaction using the fast cycling mode in a Vii7A real time PCR system:
Enzyme activation at 95°C, 2 min (1 cycle)
40 cycles of denaturation at 95°C, 1 sec and anneal/extend/acquire at 60ºC , 20 sec
Dissociation curve conditions (melt curve stage): 95°C-15 sec, 60°C-1 min, and 95°C-15 sec.
Transfer the Trizol lysate to a fresh 1.5 mL tube. NOTE: Trizol lysate can be stored at -80°C (Optional STOP step)
Add 100 µl chloroform to the Trizol lysate and vortex for 0h 0m 30s.
Centrifuge the samples at14000x g,0h 0m 0s for 0h 15m 0s at4°Cto separate aqueous, interphase and organic phases.
Transfer the top layer aqueous phase to a fresh 1.5 mL tube and add an equal volume of isopropanol; gently tap the tubes to mix. NOTE: Use a P200 pipette set at 50 µl to collect the aqueous phase. This helps to collect aqueous phase without disturbing the organic phase.
Store the tube at -20°C (Optional STOP step) or leave the tubes at Room temperature for 0h 5m 0s and centrifuge at 13000x g,0h 0m 0s for 0h 10m 0s.
Discard the supernatant using a 1 mL pipette add 0.5mL 80% (v/v) ethanol to the tube
Centrifuge at 13000x g,0h 0m 0sfor 0h 10m 0s at Room temperature.