A flow cytometry-based protocol for syngenic isolation of neurovascular unit cells from mouse and human tissues
Daniel Spitzer, Maryam I. Khel, Tim Pütz, Jenny Zinke, Xiaoxiong Jia, Kathleen Sommer, Katharina Filipski, Frits Thorsen, Thomas M. Freiman, Stefan Günther, Karl H. Plate, Patrick N. Harter, Stefan Liebner, Yvonne Reiss, Mariangela Di Tacchio, Sylvaine Guérit, Kavi Devraj























Extended
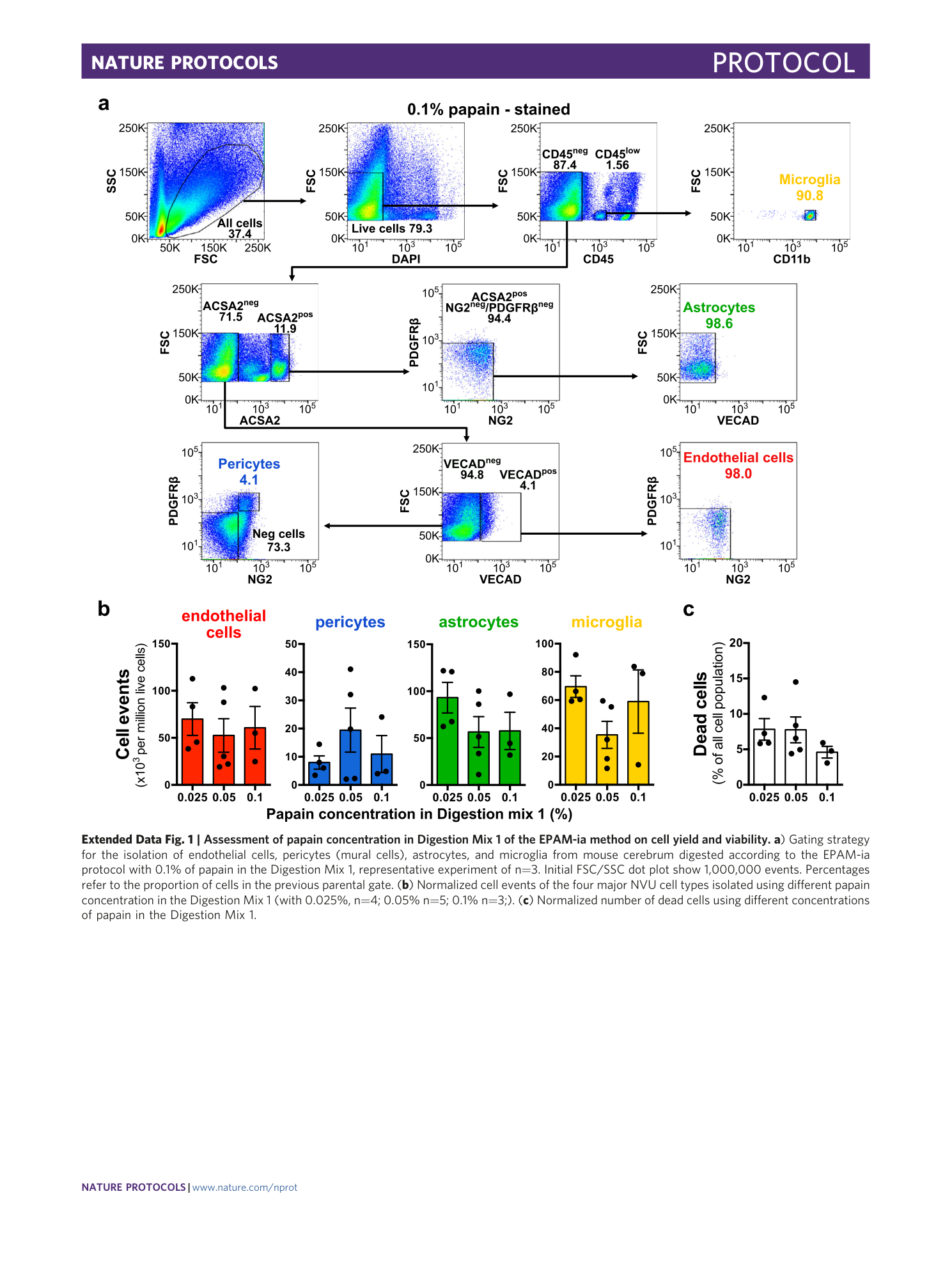
Extended Data Fig. 1 Assessment of papain concentration in Digestion Mix 1 of the EPAM-ia method on cell yield and viability.
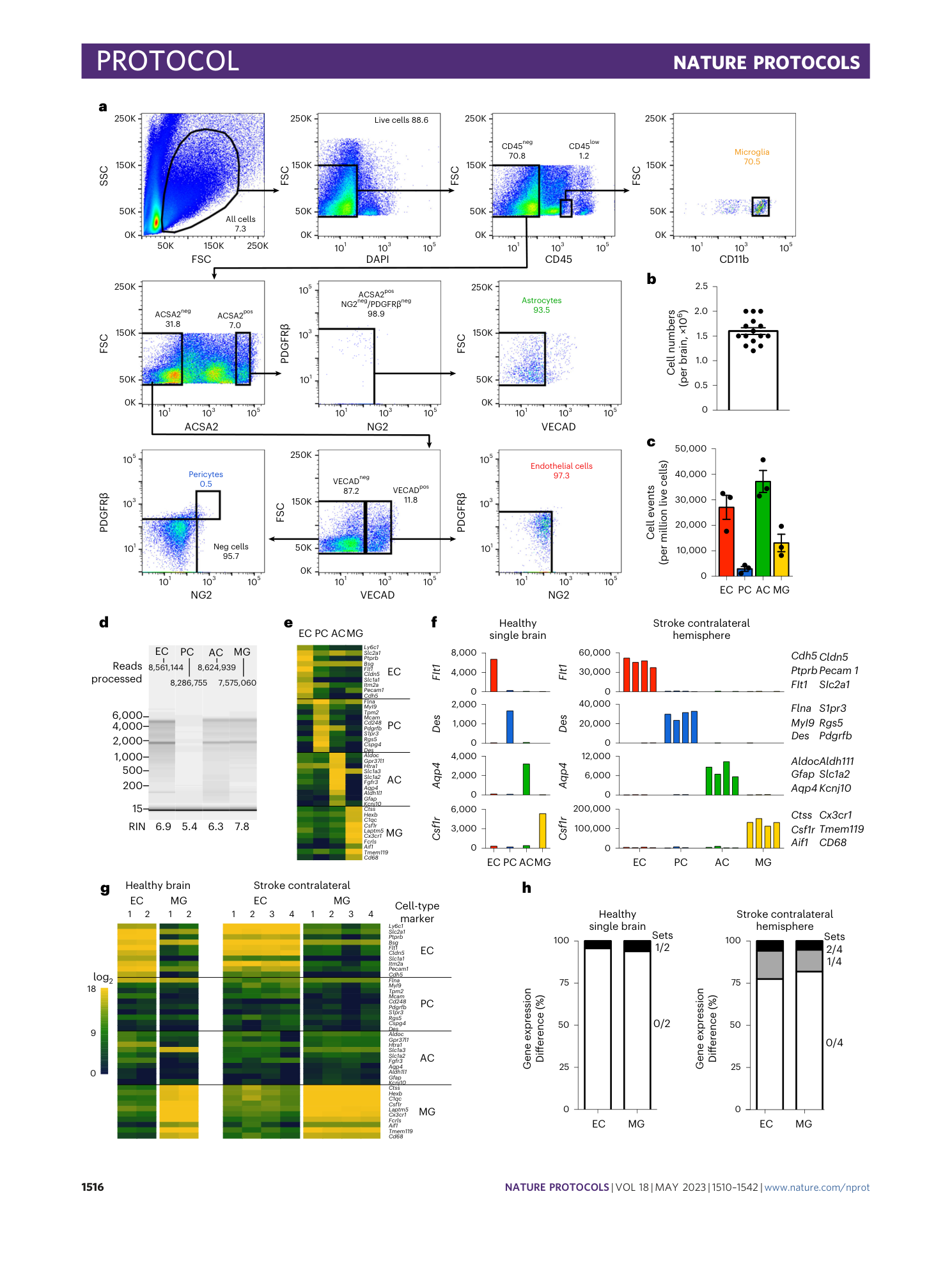
a ) Gating strategy for the isolation of endothelial cells, pericytes (mural cells), astrocytes, and microglia from mouse cerebrum digested according to the EPAM-ia protocol with 0.1% of papain in the Digestion Mix 1, representative experiment of n=3. Initial FSC/SSC dot plot show 1,000,000 events. Percentages refer to the proportion of cells in the previous parental gate. ( b ) Normalized cell events of the four major NVU cell types isolated using different papain concentration in the Digestion Mix 1 (with 0.025%, n=4; 0.05% n=5; 0.1% n=3;). ( c ) Normalized number of dead cells using different concentrations of papain in the Digestion Mix 1.
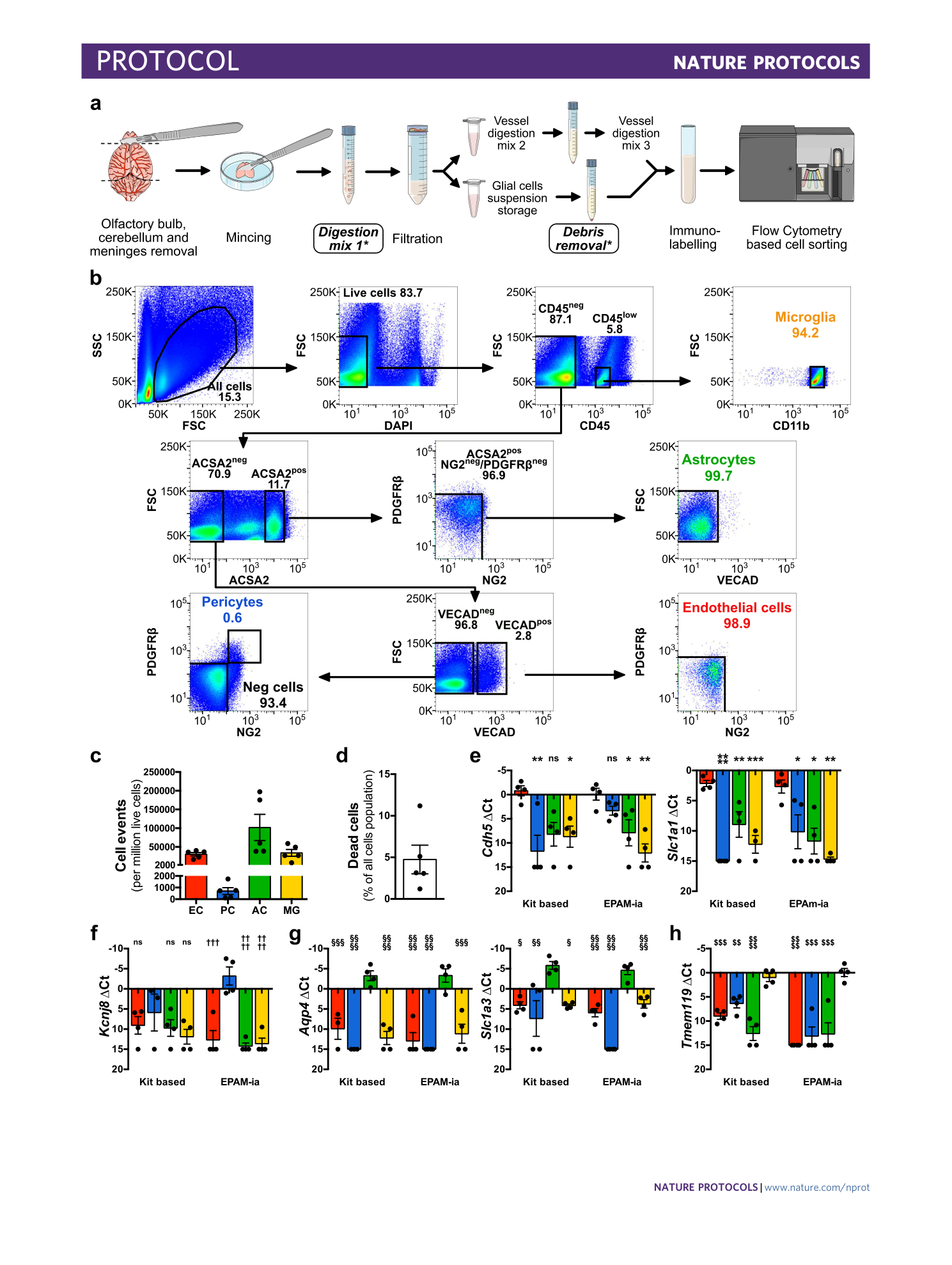
Extended Data Fig. 2 Assessment of alternative kit-based Digestion Mix 1 and debris removal solution on cell yield, viability, and purity of the isolated NVU cells.
( a ) Schematic indicating with an asterisk the two steps during the cell suspension preparation for which kit based solutions are used. ( b ) Identical gating strategy as in the fully defined EPAM-ia method is applied to obtained microglia, astrocytes, endothelial cells and pericytes (mural cells) from the same brain tissue sample with the kit based Digestion mix 1 and debris removal steps. The FSC/SSC plot shows 1,000,000 events. Percentages refer to the proportion of cells in the previous parental gate. ( c ) Normalized cell events of endothelial cells (EC), pericytes (PC), astrocytes (AC) and microglia (MG) isolated by flow cytometry after tissue dissociation using commercially available solutions for the Digestion Mix 1 and the debris removal (n=5). ( d ) Normalized number of dead cells. ( e - h ) Purity of kit-based and EPAM-ia method sorted cells is assessed by qPCR targeting cell type-specific markers of endothelial cells ( e , Cdh5 , Slca1a1 ), pericytes ( f , Kcnj8 ), astrocytes ( g , Aqp4 , Slc1a3 ), and microglia ( h , Tmem119 ). If no amplification was detected, the ∆Ct value is set at 15 by default (n=4). One way ANOVA followed by Dunnett’s multiple comparison test. *, †, § and $ indicate comparison to EC, PC, AC and MG respectively. *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001 and ns: not significant. Panels a,e – h adapted with permission from ref. [ 31 ](https://www.nature.com/articles/s41596-023-00805-y#ref-CR31 "Spitzer, D. et al. Profiling the neurovascular unit unveils detrimental effects of osteopontin on the blood–brain barrier in acute ischemic stroke. Acta Neuropathol. https://doi.org/10.1007/s00401-022-02452-1
\(2022\).") , Springer.
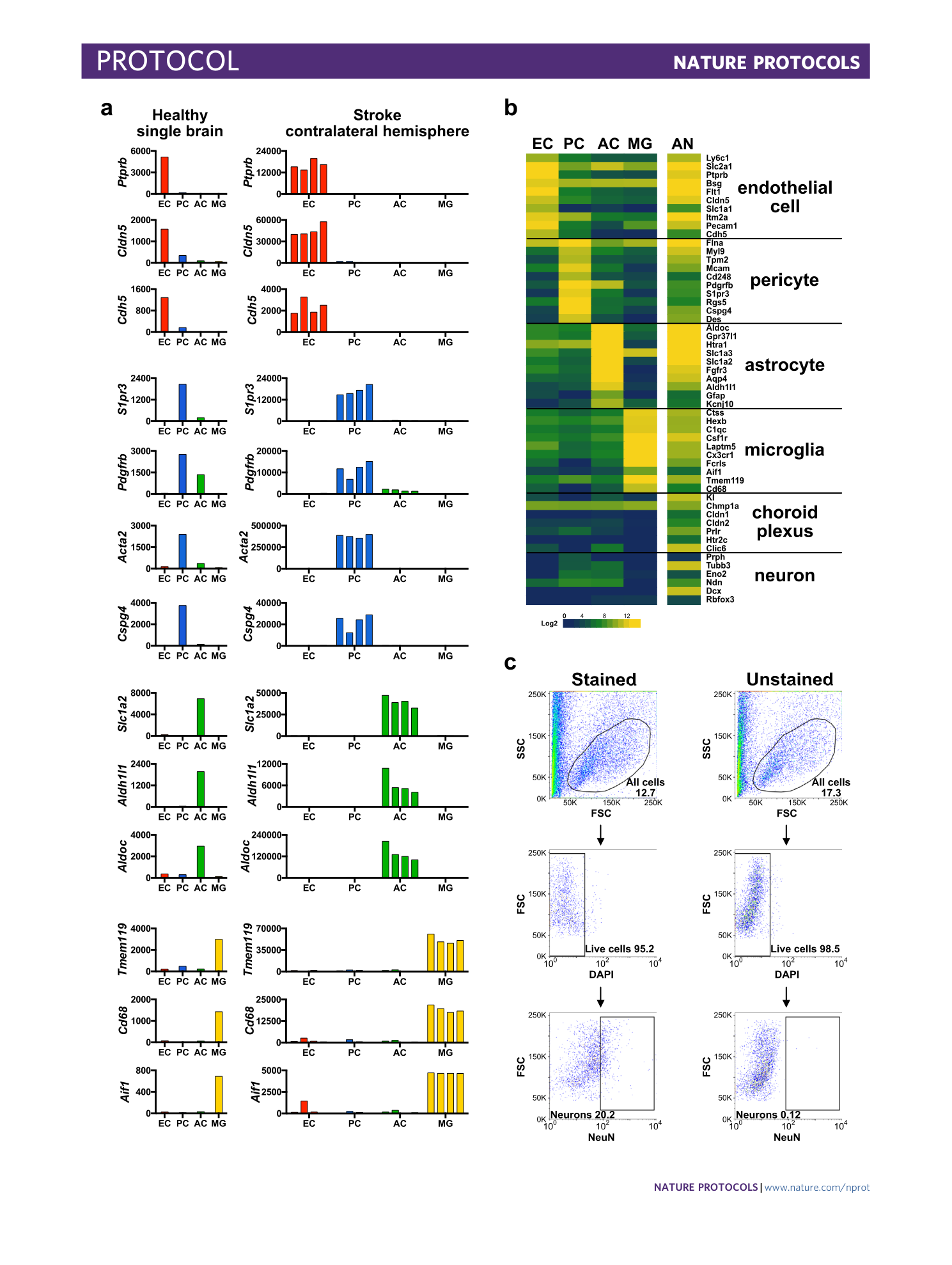
Extended Data Fig. 3 Enrichment analysis of the isolated NVU cell types and the all negative population.
( a ) Detailed transcripts reads for representative cell type markers genes of endothelial cells (EC), pericytes (PC), astrocytes (AC) and microglia (MG) in healthy single brain (n=1) or stroke contralateral hemisphere tissue (n=4, 3-4 mice/preparation). ( b ) Heatmap representing log2 expression of selected choroid plexus and neuronal cell-specific marker genes in endothelial cells, pericytes, astrocytes, microglia and all negative (AN) population isolated from a single mouse cerebrum (n=1). ( c ) FACS plots showing isolation of NeuN positive neurons by EPAM-ia method. Cells isolated by EPAM-ia method were preselected with ACSA2 magnetic beads followed by brief fixation (0.4% PFA) and permeabilization (0.1% Triton X-100) before immunostaining (n=2). Percentages refer to the proportion of cells in the previous parent gate. First SSC/FSC gate shows 200,000 events.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8.

