ANTIBIOGRAM AND PREVALENCE OF ESBL GENES IN COMMENSAL E. COLI ISOLATED FROM THE RESIDENTS OF GHANAIAN ELDERLY NURSING CARE HOMES
Idun Bright, Emmanuel Odartei Armah, Lawrencia Osae Nyarko, Mawutor Kwame Ahiabu, Agyapong Isaac, Kwarteng Freda Boampong, Oppong Mercy, Mohammed Naael, Fleischer C. N Kotey, Osei-Atweneboana Mike Yaw, Dayie Nicholas
Abstract
We
isolated E. coli i from urine samples of elderly patients and determined
the antibiogram of these pathogens. We further determined the Extended Spectrum
Beta-lactamase (ESBL) genes they harbored and their phylogenetic grouping. The
expected results were to reveal the prevalence and antibiogram of the isolated commen E. coli coli, heir carriage rate of ESBL genes, and the
phylogenetic groupings.
Steps
Isolation of Escherichia coli.
The urine samples were cultured on
Cystine-Lactose-Electrolyte-Deficient CLED agar for primary isolation.
Presumptiv E. coli li, which appeared
as yellow colonies, smooth, round, and moist after 24 hours of incubation were
kept in nutrient broth to await secondary isolations. For secondary isolation, the suspec E. coli coli isolates were cultured on
MacConkey and Blood Agar media (OXOID, Hampshire, England). The Blood agar base
was supplemented with 10% horse blood. A loop full of each sample from the
transport media was introduced on the media plate and was streaked
appropriately with a sterile inoculating loop. The media plates were
appropriately labeled and incubated at 37ºC for 24 hours. The following
biochemical tests were performed to confirm further the suspected isolates:
Indole test, Citrate test, oxidase test, and catalase test.
Biochemical Tests
Indole test
Peptone water suspensions were prepared in
a bottle according to the manufacturer’s protocol. Three to five pure isolates
were then cultured in suspensions and grown overnight. Two to three drops of
Kovac’s reagent were added to the suspension and the bottle was shaken. The formation
of a pink-colored ring that rose to the surface was observed, indicating a
positive result.
Citrate test
The Simon’s citrate agar was prepared
according to the manufacturer’s protocol. Pure isolates of the organisms on
nutrient agar were inoculated into the citrate agar and incubated for 24
hrs. The citrate agar was green before
inoculation. There was no color change beca E. coli coli is citrate-negative.
Oxidase test
A
drop of oxidase reagent, which contains tetramethyl-p-phenylenediamine was
placed on a pure colony of th E. coli li. No color change occurred indicatin E. coli oli exhibits a negative oxidase reaction.
Catalase test
A
few drops of hydrogen peroxide were added to pure colonies of E. coli i. There was an immediate release
of oxygen bubbles, indicating a positive reaction.
Antibiogram of E.coli isolates; Kirby-Bauer disc
diffusion test
The Kirby-Bauer antimicrobial sensitivity
test method was used to determine the antibiogram of the E. coli i isolates (Bauer et al., 1966). Ten
antimicrobial drugs were used. These were imipenem
(IPM, 10 µg), ertapenem (ERT, 10 µg), aztreonam (AZM, 15 µg), cefepime (FEP, 30
µg), nitrofurantoin (F, 50 µg), cefuroxime (CXM, 10 µg), gentamycin (CN, 10
µg), amikacin (AK, 30 µg), ciprofloxacin (CIP, 5 µg) and levofloxacin (LEV, 10
µg).
Mueller-Hinton agar was prepared according
to the manufacturer’s protocol. The organisms were cultured on nutrient agar
overnight. Between 4 and 5 isolated colonies of the organisms were suspended in
about 2 ml of sterile saline by use of inoculating loop. The saline tube was
vortexed to create a smooth suspension. The turbidity of the suspension was
adjusted to a 0.5 McFarland standard. 200 ml of the suspension was introduced onto
the Mueller-Hinton plate. A sterile glass spreader was used to spread the
organisms on the plate. The surface of the plate was allowed to dry for 5 minutes
before the antibiotic discs were placed on them. Sterile forceps were used to
remove the antibiotic discs from the dispensers. After placing the discs on the
agar, each disc was gently touched with the inoculating loop to ensure their
contact with the agar surface. The plates were then incubated upside down for
24 hours at 37ºC.
PCR Screening for ESBL Genes
The DNAs
of the 41 E. coli i isolates were
extracted using a zymogen extraction kit, based on the manufacturer’s protocol.
The isolates were then screened to determine the types of beta-lactamas bla bla) genes they harbored. A total of forty-o E. coli coli isolates were screened for
the presence o bla 0 bla genes. The
protocols employed by Kiiru and colleagues (Kiiru et al.,
- were used with slight modifications. The
reactions were carried out in a 10 µl reaction volume. This consists of 5 µl of
2X
SYBR green master mix, 0.2 µl
each of the primer sequence, 2.6 µl of the Nuclease free water, and 2 µl of the
DNA template. The primer concentration was 0.2M. The PCR cycle
conditions were as follows: 3 mins of initial denaturation at 94 ºC, (94 ºC of
denaturation for 30 seconds, annealing for 30 seconds, elongation at 68 ºC for
30 seconds) x 30 cycles, and final elongation at 68 ºC for 10 minutes. The
annealing temperatures were different for the different primers. The primer sequence
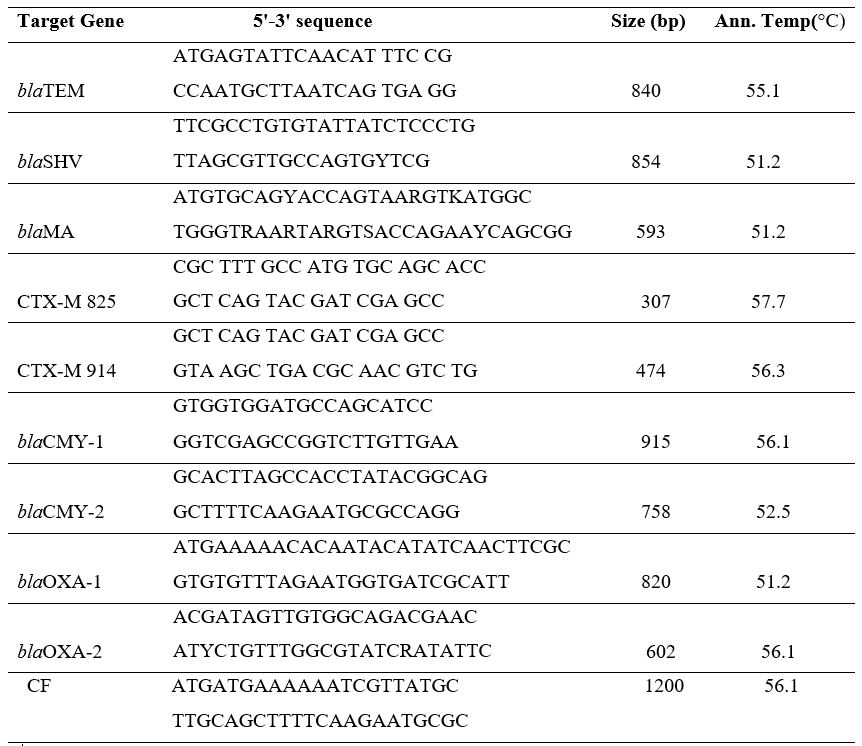
and the annealing temperatures are listed in Table 1
Table 1: Primer sequences of the ESBL genes, amplicon sizes, and annealing temperatures s
Phylogenetic Grouping by PCR
As described by Clermont et al. , 2000, the phylogenetic grouping
of the isolated E. coli i was
determined. The positiv E. coli i strains
were investigated for various genes that would determine their phylogenetic
grouping by multiplex PCR (Clermont et al., 2000). The procedures were
performed in a 10 µl reaction mixture. The reaction included 5 µl of 2X SYBR
green master mix, 0.2 µl each oligonucleotide primer, 2.6 µl of nuclease-free
water, and 2 µl of template DNA. Primer concentration was 0.2 M. Conditions of
the reaction mixtures were 3 mins at 94 ºC initial denaturation, (94 ºC of
denaturation for 30 seconds, annealing at 59.2 °C for 30 seconds, elongation at
68 ºC for 3 minutes) x 30 cycles, and final elongation at 68 ºC for 10 minutes.
The marker-specific primer sequences and their amplicon sizes are listed in
Table 2. Based on the presence or absence of spec chuA, yjaA, TspE4.C2, , T arpA C2 and arpA), the isolates were clustered in group A,
, B2, or D. Both group B1 and group chuA acks the chuAgene. However, group B1
TspE4.C2 the TspE4.C2 gene, which is abse t in chuA up A. The chuA
gene is present in both B2 and D isolates. The difference between the two is
t yjA roup B2 has yjA genes while group D lacks it
Table 1: Primer sequences of the ESBL genes, amplicon sizes, and annealing temperatures s
SKS