16S Gene PCR Amplification and Sanger Sequencing
Carlos Carlos Goller
Abstract
After genomic DNA (gDNA) has been isolated and quantified, it can be used for many downstream applications. While we will use our genomic DNA samples for whole genome sequencing using Nanopore sequencing technology, we can also use the gDNA to amplify one gene for Sanger sequencing. These different sequencing methods contrast one another:
Table. Comparison of Nanopore and Sanger DNA Sequencing.
| A | B |
|---|---|
| Nanopore Sequencing | Sanger Sequencing |
| High Throughput (many genes at once) | Low Throughput (one gene at a time) |
| Sequences gDNA directly | Sequences PCR amplified DNA |
| Ion current base calling | Fluorophore base calling |
PCR amplification of 16S
We will amplify the bacterial 16S gene, which encodes a subunit of ribosomes. Biologists use this gene to compare evolutionary relationships by sequence similarities and differences to organize biological taxonomies. Polymerase Chain Reaction (PCR) enables scientists to amplify many copies of a gene, and we will target 16S for amplification.
Steps
Introduction
#尊敬的用户,由于网络监管政策的限制,部分内容暂时无法在本网站直接浏览。我们已经为您准备了相关原始数据和链接,感谢您的理解与支持。
https://lh7-us.googleusercontent.com/mpzdksVN3LG0gvqZPp2tdPiBgdxqzJ6qac9sS-erd-0jT-xdGtTrCCYF0LvLzq2zzVUOwFxNjf9CPP3bOVMU-OFKuJupdQ5nrTllaxWsL1la2T7tYJzMGNU2JcHfQFs90BHLGse-wOrdBMc7Lf0PDXM
PCR Cycles Diagram indicating the three main steps: denaturation at 95-96°C, annealing at 68°C, and elongation at 72°C. Created with BioRender.com
You can learn about this reaction by watching the video “Polymerase Chain Reaction.”
After completing this lab, you will gain the following lab skills:
- Lab safety and proper personal protective equipment (PPE)
- Setup of two PCR reactions.
- Proper use of a thermocycler for PCR.
PCR Setup
Note Before You Begin:
Review the protocols and figures below to learn how the 16S PCR will help us amplify regions of this gene.
| A | B | C |
|---|---|---|
| Reagent | Volume | Final Concentration |
| DNA polymerase master mix (contains proof-reading DNA polymerase, dNTPs, buffer, Mg2+)NEB Q5 Hot Start High-Fidelity 2X Master Mix. | 25 µL | 1X |
| 10 µM Forward Primer | 2.50 µL | 0.5 µM |
| 10 µM Reverse Primer | 2.50 µL | 0.5 µM |
| Nuclease-free water | 17.5 µL | - |
| gDNA (<100 ng/µL) | 2.5 µL | <1,000 ng |
| FINAL REACTION | 50 µL | - |
PCR Table. Reagents and volumes needed for 16S PCR.
For each gDNA sample, set up two reactions :
- one LONG reaction with primers 27fwd + 1492rev
- one SHORT reaction with 515fwd + 1492rev
We will use the
| A | B |
|---|---|
| Primer Name | Primer sequence 5’ → 3’ |
| 27fwd | AGA GTT TGA TCM TGG CTC AG |
| 515fwd | GTG CCA GCM GCC GCG GTA A |
| 1492rev | CGG TTA CCT TGT TAC GAC TT |
Primer Table. Primer names and sequences for the amplification of 16S.
#尊敬的用户,由于网络监管政策的限制,部分内容暂时无法在本网站直接浏览。我们已经为您准备了相关原始数据和链接,感谢您的理解与支持。
https://lh7-us.googleusercontent.com/uT0h1n-UZzkWtUenKSE6HkquvBOLQNAAotodjeDSFqR7S4ZNonTP7SCQCoQu7iTv_UjP4vzaQJqFPqO3zxCpUUQHV-HaL3HL3mEBAxIfFznxsO7k15TjeLsoIAHYmrqfcZkWVjjt12Tx4VsfS6ANOac
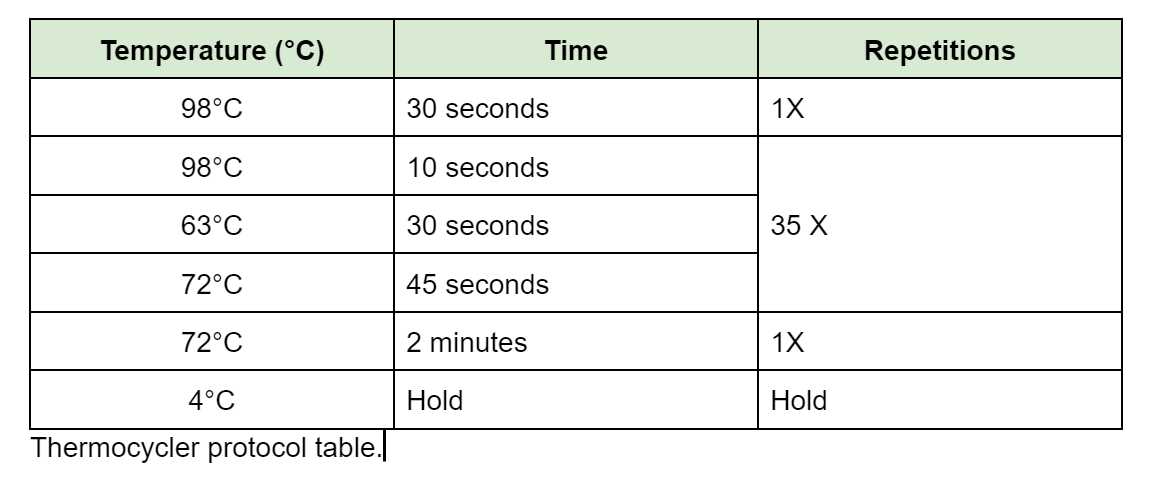
PCR Clean-up
To remove the reagents from the PCR and isolate pure DNA for Sanger sequencing, we need to "clean up" our amplified DNA samples. In BIT 295, we use reagents from the
Add all 50µL of the PCR reaction to five volumes (250µL) PB buffer
Mix and transfer to pink spin column
Sample Centrifuge at 10.000rpm
Remove flow through
Add 750 µL PE to wash the column. Incubate at RT for 1 minute.
Centrifuge at 10.000rpm
Remove the spin column from the collection tube and add to a new wash tube
Centrifuge at 10.000rpm to remove all ethanol wash
Remove the spin column from the collection tube and add itto a new collection tube
Add 50µL EB to elute. Incubate at RT for 1 minute.
2µL Centrifuge at 10.000rpm
Remove the spin column and throw away
Quantify 2 µl using the Implen that has been blanked with the elution buffer (EB).
DNA can be stored at 4°C or on ice while being used and should be stored long-term at -20°C
Sanger Sequencing Sample Preparations
For Sanger sequencing, start by sending out of ~100 ng/µL 20µL of ~100 ng/µL DNA of your LONG amplicon for three sequencing reactions with all three primers 27fwd, 515 fwd, and 1492rev. Prepare 10µL a 1:10 dilution of your 100 µM primers.
If you do not get clear chromatograms to interpret the sequence, next send out your SHORT amplicon for Sanger sequencing with only primers 515fwd and 1992rev. Prepare 10µL a 1:10 dilution of your 100 µM primers.
16S rRNA Sequencing: A PCR-based Technique to Identify Bacterial Species
Adapted from: Identification of unknown bacterial isolates using Sanger sequencing of the 16S rRNA gene | CHMI services.

